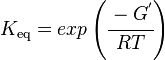Limonene Synthase
You can go back to main page of the kinetic model here.
In this model, LIMS is modelled in Escherichia coli and this model is replicating the bacterial system in vivo. As such, in vivo-like conditions such as pH of 7.5 and temperature of 30°C in E. coli is set as the ideal conditions when assigning weights to its parameter. Limonene synthase catalyses the formation of limonene and pyrophosphate from one molecule of geranyl diphosphate (GPP). The following equations show LIMS’s reaction stoichiometry and its corresponding reaction rate using the Michaelis-Menten rate law:
Contents
Equation Rate
The reversible Michaelis-Menten equation to model the dynamic changes of LimSynth is:
where :
| Parameter | Description | Units |
|---|---|---|
| VLimSynth | Reaction rate for Limonene Synthase | |
| Vmaxforward | Maximum reaction rate towards the production of limonene | ref |
| KmGPP | Michaelis-Menten constant for GPP | mM |
| KmLimonene | Michaelis-Menten constant for Limonene | mM |
| KmPP | Michaelis-Menten constant for PP | mM |
| Keq | Equilibrium constant | |
| [GPP] | GPP concentration | mM |
| [Limonene] | Limonene concentration | mM |
| [PP] | PP concentration | mM |
Metabolite Background Information
Long metabolite names are abbreviated in the model for clarity and standard identification purposes.
| Metabolite | Abbreviation | Chemical Formula | Molar mass (g/mol) | ChEBI | ChEMBL | PubChem |
|---|---|---|---|---|---|---|
| geranyl diphosphate | GPP | C10H20O7P2 | 314.209 | 17211 | 41432 | 445995 |
| (-)-4S-limonene | Limonene | C10H16 | 136.24 | 15384 | 449062 | 22311 or 439250 |
| diphosphate | PP | O7P2 | 173.94 | 644102 | ||
| limonene synthase | LimSynth | 70.03 kDa [1], 72.4 kDa [2] ; 60kDa [3]; 56 kDa [4] |
Parameterisation
Calculating the Equilibrium Constant
Unlike the kinetic parameter values, thermodynamic parameter values such as for equilibrium constant (Keq) are not easily found in literature reports. However, Keq can be calculated from Gibbs Free Energy (ΔG°) using the following equation:
where;
| Keq | Equilibrium constant |
| -ΔG° | Gibbs free energy change (kcal/mol) |
| R | Gas constant (0.0019859 kcal/K/mol) |
| T | Absolute temperature (298 K) |
Gibbs free energy values for LIMS are obtained from MetaCyc (EC 4.2.3.16) is -28.049988 kcal/mol [5] and Equilibrator [1]. Table 2 summarizes the ΔrG° values found for LIMS and the calculated Keq. These Keq values are given an arbitrary equal weight of 1 as the ΔrG° values obtained were calculated from a group contribution method and do not have any measurement conditions that would allow us to assess the Keq values according to the weighting scheme set out in (insert link here).
| ΔrG°(kcal/mol) | Keq | Error (±) | Source | Weight |
|---|---|---|---|---|
| -28.050 | 3.843E+20 | N/A | MetaCyc | 1 |
| -42.161 ± 2.844 | 8.711E+30 | 5.876E+29 | Equilibrator | 1 |
Kinetic Parameter Values
Assessing kinetic parameter values. Values for kinetic parameters such as for KmGPP and Kcatforward can be obtained from extensive literature searches. Our findings from literature searches of these parameters are summarised in Table 1. In addition to the parameter values, the measurement conditions such as the pH, temperature and gene source used for the kinetic evaluations are also taken into consideration. Then, in order to assess the plausibility of the parameter values collected, weights are assigned according to different weighting categories as described in Table 2. Final weight for each parameter value is calculated by taking each resulting weight from each category and multiplying it. For example, Lücker et. al. isolated limonene synthase cDNA from lemon (Citrus limon) and functionally expressed them in Escherichia coli. Enzyme assays were conducted at pH 7.0 and 30°C, and the resulting measured KmGPP is at 0.7 µM. This parameter value (KmGPP) is assigned a total weight of 32 (1x1x4x2x4=32) after assessing that:- a) this value was measured in vitro (weight=1), b) limonene synthase isolated was from lemon which is unrelated to E. coli (weight=1), c) limonene synthase measured was from the same EC (EC 4.2.3.16, weight=4), d) measurement pH was within ± 0.5 from the ideal pH of 7.5 (weight=2), and finally e) measurement temperature was at the ideal 30°C (weight=4). The breakdown of weights assigned to each kinetic parameter value found for limonene synthase is shown in Table below.
A more detailed descriptions of the values listed above can be found HERE , where I've linked and highlighted where these data came from.
| Name | Value | Units | Remarks-References | |
|---|---|---|---|---|
| KmGPP | 6.8 | µM | This study has isolated and characterised a (-)-limonene synthase recombinant encoded by Cannabis sativa L. cv. ‘Skunk’ trichome mRNA. pH optimum was determined at pH 6.5 and a temperature optimum at 40°C. (Günnewich,, N., 2007) | |
| Kcatforward | 0.082 | s-1 | ||
| KmGPP | 47.4 ± 3.8 | µM | In this study,limonene synthase were cloned from lavender (Lavandula angustifolia) leaves and flowers. Temperature was determined at 30°C and pH at 7.0. (Landmann,C 2007) | |
| Kcatforward | 0.012 | s-1 | ||
| KmGPP | 6.7 | µM | This study reports the kinetic evaluation the native form of LIMS from Mentha spicata. Reaction mixtures were determined at pH 7.0 and temperature of 30°C. Kinetic constants are averaged data sets of triplicate analyses (n=3). (Williams 1998) | |
| Kcatforward | 0.024 | s-1 | ||
| Kcatforward | 0.3 | s-1 | In this study, the authors have isolated and purified limonene synthase from both peppermint (Mentha x piperita) and spearmint (Mentha spicata). Enzyme activity was determined at pH 7.0m and temperature 30°C. (Alonso 1992) | |
| KmGPP | 1.25 | µM | In this study,
limonene synthase is characterised and extracted from liverwort (Ricciocarpus natans). The enzyme assay was conducted at pH 6.5 and at temperature of 32°C. (Adam 1996) |
|
| KmGPP | 0.7 | µM | This study isolated limonene synthase cDNA from lemon (Citrus limon) and functionally expressed them in Escherichia coli. Enzyme assays were
conducted at pH 7.0 and 30°C. (Lucker 2002) |
|
| KmGPP | 1.8
± 0.3 |
µM | Limonene synthase from purified preparation of glandular trichome seceratory cell clusters from peppermint (Mentha x piperita). Enzyme assays were carried out in buffer pH 7.0, and incubated at 30°C. Data reported are the averages of three independent assays (n=3). (Rajaonarivony 1992) |
Vmax values
| Vmax | Unit | Directionality | Organism | Organism | References |
|---|---|---|---|---|---|
| 0.08 | µmol/min/mg | forward | Cannabis sativa L. | [1] | |
| 0.12 ± 0.01 | µmol/min/mg | forward | Cannabis sativa | [1] | |
| 0.4748 | µmol/min/mg | forward | Citrus limon | References | |
| 0.53 | µM/min | forward | Citrus sinensis | 200µM GPP, 20°C, [E]: 0.07µM, pH7.5 | [6] |
| 0.64 | µM/min | forward | Citrus sinensis | 200µM GPP, 37°C, [E]: 0.26µM, pH7.5 | [6] |
| 19 | µmol/h/mg | forward | Mentha x piperita & Mentha spicata | Maximum specific activity, 1-10µg protein per mixture, pH &.0, 30°C | References |
Kcat values
| Parameter | Direction | Substrate | Value | Unit | Method | M_w | Organism | O_w | Expression_vector | E_w | Enzyme | En_w | pH | P_w | Temperature | T_w | Total_weight | Description | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kcat | Forward | GPP | 0.0004 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp, expressed in E.coli and truncated (R59), pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0004 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli , truncated R58P59, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0004 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli , truncated R58A59, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0004 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli , S60, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.002 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli preprotein, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0024 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli wt, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0034 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli E57, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0036 | 1/s | in vitro | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli Q54, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0037 | 1/s | in vitro | 2 | Unrelated (+0) | 0 | E.coli | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli R58, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.02 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli recombinant preprotein , pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.04 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | close range (+2) | 2 | 37°C (+4) | 4 | 256 | Citrus sinensis, expressed in E.coli 200µM GPP, 37°C, [E]: 0.26µM, pH7.5 | Entova2013 |
| Kcat | Forward | GPP | 0.082 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | close range (+2) | 2 | 36°C - 40°C (+2) | 2 | 128 | Cannabis sativa L., expressed in E.coli,CsTPS1, pH6.5, 40°C, 1.25 mg [LimSynth] per 500ml assay mixture, 10mM [GPP] | Gunnewich2008 |
| Kcat | Forward | GPP | 0.09 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | close range (+2) | 2 | 36°C - 40°C (+2) | 2 | 128 | Cannabis sativa L., expressed in E.coli,Km of 6.25 ± 0.41 µM, pH 6.5, 40°C, [GPP] 0.5 - 25.0 µM used for kinetic parameter determination, kcat 0.09 /s | Gunnewich2008 |
| Kcat | Forward | GPP | 0.13 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | close range (+2) | 2 | 36°C - 40°C (+2) | 2 | 128 | Cannabis sativa L., expressed in E.coli,Km of 6.25 ± 0.41 µM, pH 6.5, 40°C, [GPP] 0.5 - 25.0 µM used for kinetic parameter determination, kcat 0.09 /s | Gunnewich2008 |
| Kcat | Forward | GPP | 0.13 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | close range (+2) | 2 | different temperature (+0) | 0 | 128 | Citrus sinensis, expressed in E.coli 200µM GPP, 20°C, [E]: 0.07 mM, pH7.5 | Entova2013 |
| Kcat | Forward | GPP | 0.3 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Mentha piperita & Mentha spicata, expressed in E.coli , pH7.0, 30°C, enzyme assay, 1 -10 mg protein per mixture | Alonso1992 |
| Kcat | Forward | GPP | 0.000039 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 512 | Citrus sinensis, expressed in E.coli40mM GPP, 37°C, [E]: 0.3 mM, pH7.0 (check units) | Olsen2011 |
Extracting Information from Limonene Production Rates
The production rates would reflect on the flux for this enzyme, and this would provide provide the insights on the Vmax of this enzyme.
| Amount produced (mg/L) | Time (H) | Organism | Description | Reaction Flux (µM/s) |
|---|---|---|---|---|
| 5 | 24 | Escherichia coli | Possible reason for the low limonene production might due to the insufficient supply of IPP and DMAPP [7]. | 0.0255 |
| 335 | 48 | Escherichia coli | Engineered E.coli in which heterologous MVA pathway was installed [8]. | 0.8537 |
| 35.8 | 48 | Escherichia coli | E.coli was engineered to express GPPS, LS, DXS, and IDI [9] . | 0.0912 |
| 4.87 | 48 | Escherichia coli | This was the initial titer. The study established a limonene biosynthesis pathway in E.coli using four different polycistronic operons based on 3 vectors with varied expression strength [9]. | 0.0124 |
| 17.4 | 48 | Escherichia coli | Using a plasmid with DXS and IDI over expressed [9]. | 0.0445 |
| 430 | 72 | Escherichia coli | [8] | 0.7306 |
Parameter estimation
This section can be found HERE
Simulations
Simulations performed can be found HERE.
References
- ↑ 1.0 1.1 1.2 Gunnewich, N. 2008. "Expression and characterization of terpene synthases from Cannabis sativa L. and Salvia sclarea L. Doctoral thesis. Cite error: Invalid
<ref>tag; name "Gunnewich2008" defined multiple times with different content - ↑ Turner,G. et. al.1999. "Limonene synthase, the enzyme responsible for monoterpene biosynthesis in peppermint, is localized to leucoplasts of oil gland secretory cells", Plant Physiology 120(3): 879-886
- ↑ Maruyama, T. et. al. 2002. "Molecular cloning, functional expression and characterization of d-Limonene synthase from Agastache rugosa" Biol. Pharm. Bull. 25(5): 661-665
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedAlonso1992 - ↑ Latendresse M. (2013). "Computing Gibbs Free Energy of Compounds and Reactions in MetaCyc."
- ↑ 6.0 6.1 Cite error: Invalid
<ref>tag; no text was provided for refs namedEntova2013 - ↑ Carter, Ora A. et. al.2013. "Monoterpene biosynthesis pathway construction in Escherichia coli",Phytochemistry, 64:425–433, 2003.
- ↑ 8.0 8.1 Alonso-Gutierez et. al. 2013. "Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production", Metabolic Engineering, 19:33-41 Cite error: Invalid
<ref>tag; name "AlonsoGutierez2013" defined multiple times with different content - ↑ 9.0 9.1 9.2 Du et. al. 2014. "Enhanced limonene production by optimizing the expression of limonene biosynthesis and MEP pathway genes in E.coli", Bioprocessing and Bioprocessing, 1:10
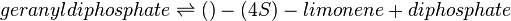
![V_\mathrm{LimSynth} = Vmax_\mathrm{forward} * \cfrac {\cfrac{[GPP]}{Km_\mathrm{GPP}} * \left ( 1 - \cfrac {[Limonene]*[PP]}{[GPP]*K_\mathrm{eq}} \right )}{1 + \cfrac {[GPP]}{Km_\mathrm{GPP}} + \cfrac {[Limonene]}{Km_\mathrm{Limonene}} + \cfrac {[PP]}{Km_\mathrm{PP}} + \cfrac {[Limonene]*[PP]}{Km_\mathrm{Limonene}*Km_\mathrm{PP}}}](/wiki/images/math/6/9/8/698f16353b522504a844aae5c37bdac9.png)