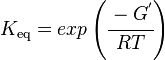Limonene Synthase
You can go back to main page of the kinetic model here.
In this model, LIMS is modelled in Escherichia coli and this model is replicating the bacterial system in vivo. As such, in vivo-like conditions such as pH of 7.5 and temperature of 30°C in E. coli is set as the ideal conditions when assigning weights to its parameter. Limonene synthase catalyses the formation of limonene and pyrophosphate from one molecule of geranyl diphosphate (GPP). The following equations show LIMS’s reaction stoichiometry and its corresponding reaction rate using the Michaelis-Menten rate law:
Contents
Equation Rate
The reversible Michaelis-Menten equation to model the dynamic changes of LimSynth is:
where :
| Parameter | Description | Units |
|---|---|---|
| VLimSynth | Reaction rate for Limonene Synthase | |
| Vmaxforward | Maximum reaction rate towards the production of limonene | ref |
| KmGPP | Michaelis-Menten constant for GPP | mM |
| KmLimonene | Michaelis-Menten constant for Limonene | mM |
| KmPP | Michaelis-Menten constant for PP | mM |
| Keq | Equilibrium constant | |
| [GPP] | GPP concentration | mM |
| [Limonene] | Limonene concentration | mM |
| [PP] | PP concentration | mM |
Metabolite Background Information
Long metabolite names are abbreviated in the model for clarity and standard identification purposes.
| Metabolite | Abbreviation | Chemical Formula | Molar mass (g/mol) | ChEBI | ChEMBL | PubChem |
|---|---|---|---|---|---|---|
| geranyl diphosphate | GPP | C10H20O7P2 | 314.209 | 17211 | 41432 | 445995 |
| (-)-4S-limonene | Limonene | C10H16 | 136.24 | 15384 | 449062 | 22311 or 439250 |
| diphosphate | PP | O7P2 | 173.94 | 644102 | ||
| limonene synthase | LimSynth | 70.03 kDa [1], 72.4 kDa [2] ; 60kDa [3]; 56 kDa [4] |
Parameterisation
Calculating the Equilibrium Constant
Unlike the kinetic parameter values, thermodynamic parameter values such as for equilibrium constant (Keq) are not easily found in literature reports. However, Keq can be calculated from Gibbs Free Energy (ΔG°) using the following equation:
where;
| Keq | Equilibrium constant |
| -ΔG° | Gibbs free energy change (kcal/mol) |
| R | Gas constant (0.0019859 kcal/K/mol) |
| T | Absolute temperature (298 K) |
Gibbs free energy values for LIMS are obtained from MetaCyc (EC 4.2.3.16) is -28.049988 kcal/mol [5] and Equilibrator [1]. Table 2 summarizes the ΔrG° values found for LIMS and the calculated Keq. These Keq values are given an arbitrary equal weight of 1 as the ΔrG° values obtained were calculated from a group contribution method and do not have any measurement conditions that would allow us to assess the Keq values according to the weighting scheme set out in (insert link here).
| ΔrG°(kcal/mol) | Keq | Error (±) | Source | Weight | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -28.050 | 3.843E+20 | N/A | MetaCyc | 1 | |||||||||||||||
| -42.161 ± 2.844 | 8.711E+30 | 5.876E+29 | Equilibrator | 1 |
| Concentration | Unit | Substrate / Product | Directionality | Organism | Method notes | References |
|---|---|---|---|---|---|---|
| 10 | µM | GPP | forward | Citrus sinensis | enzyme activity assay, pH 7, 37°C, with 0.3µM enzyme | [6] |
| 40 | µM | GPP | forward | Citrus sinensis | enzyme activity assay, pH 7, 37°C, with 0.3µM enzyme | [6] |
| 0.3 | µM | Limonene synthase | - | Citrus sinensis | enzyme activity assay, pH 7, 37°C, with 10-40µM GPP | [6] |
| 25 | µM | GPP | forward | E. coli | [7] | |
| 10 | µM | GPP | forward | Cannabis sativa L. | [8] | |
| 10 | µM | GPP | forward | Mentha sp. | [4] | |
| 2 | µM | GPP | forward | Citrus limon | [9] | |
| 150 | µg | Limonene synthase | - | E. coli | [7] | |
| 0.07 | µM | Limonene synthase | - | Citrus sinensis | with 200µM GPP as substrate, at 20°C, pH7.5 | [10] |
| 0.26 | µM | Limonene synthase | - | Citrus sinensis | with 200µM GPP, at 37°C, pH7.5 | [10] |
| 200 | µM | GPP | forward | Citrus sinensis | used for Limonene synthase activity assay at 20°C and 37°C, pH 7.5 | [10] |
| 100 | µM | GPP | - | Citrus limon | [9] | |
| 0.1 | µM | GPP | - | Citrus limon | [9] |
Km Values
| Parameter | Direction | Substrate | Value | Unit | Method | M_w | Organism | O_w | Expression_vector | E_w | Enzyme | En_w | pH | P_w | Temperature | T_w | Total weight | Description | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km | Forward | GPP | 0.0067 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Native form, protein from Mentha sp. expressed in E. coli. | [7] |
| Km | Forward | GPP | 0.7 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Citrus limon. Range of 1-100 mM [GPP] was used to determine Km of LimSynth, pH7.0 , 30°C, expressed in E.coli | [9] |
| Km | Forward | GPP | 1.25 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 512 | 5mM [GPP], 32°C, pH7, 10-20 mg protein used for enzyme assay, Ricciocarpos natans | [11] |
| Km | Forward | GPP | 1.8 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | From Mentha spicata and Mentha x piperita, pH7.0, 30°C, enzyme assay | [4] |
| Km | Forward | GPP | 6 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 512 | Protein from Mentha sp., expressed in E.coli, pH7.0, 1-50 mg [E], 25 mM [GPP], limonene produced 94%, 2% myrcene, pinene | [7] |
| Km | Forward | GPP | 6.25 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | Phylogenetically related (+2) | 2 | Identical (+4) | 4 | close range (+2) | 2 | 36°C - 40°C (+2) | 2 | 64 | Protein from Cannabis sativa, expressed in bacteria? Km of 6.25 ± 0.41 mM, pH 6.5, 40°C, [GPP] 0.5 - 25.0 mM used for kinetic parameter determination, kcat 0.09 /s | [1] |
| Km | Forward | GPP | 8.6 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 512 | protein from Mentha sp., expressed in E.coli, pH7.0, 1-50 mg [E], 25 mM [GPP], limonene produced 94%, 2% myrcene, pinene | [7] |
| Km | Forward | GPP | 11.76 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | Phylogenetically related (+2) | 2 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 256 | protein from Cannabis sativa, expressed in bacterial host. Km of 11.76 ± 2.45, pH 7.0, [GPP] 0.5 - 25.0 microM used for kinetic parameter determination. | [1] |
| Km | Forward | GPP | 12.6 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 512 | Protein from Mentha sp., expressed in E.coli. pH7.0, 1-50 mg [E], 25 mM [GPP], limonene produced 94%, 2% myrcene, pinene | [7] |
| Km | Forward | GPP | 14.9 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 512 | recombinant preprotein,Protein from Mentha sp., expressed in E.coli. pH7.0, 1-50 mg [E], 25 mM [GPP], limonene produced 94%, 2% myrcene, pinene | [7] |
| Km | Forward | GPP | 16 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 512 | recombinant preprotein,Protein from Mentha sp., expressed in E.coli. pH7.0, 1-50 mg [E], 25 mM [GPP], limonene produced 94%, 2% myrcene, pinene | [7] |
| Km | Forward | GPP | 18.8 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 512 | recombinant preprotein,Protein from Mentha sp., expressed in E.coli. pH7.0, 1-50 mg [E], 25 mM [GPP], limonene produced 94%, 2% myrcene, pinene | [7] |
| Km | Forward | GPP | 27.5 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 512 | recombinant preprotein,Protein from Mentha sp., expressed in E.coli. pH7.0, 1-50 mg [E], 25 mM [GPP], limonene produced 94%, 2% myrcene, pinene | [7] |
| Km | Forward | GPP | 29.9 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 512 | recombinant preprotein,Protein from Mentha sp., expressed in E.coli. pH7.0, 1-50 mg [E], 25 mM [GPP], limonene produced 94%, 2% myrcene, pinene | [7] |
| Km | Forward | GPP | 32.4 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 512 | recombinant preprotein,Protein from Mentha sp., expressed in E.coli. pH7.0, 1-50 mg [E], 25 mM [GPP], limonene produced 94%, 2% myrcene, pinene | [7] |
| Km | Forward | GPP | 130 | µM | in vitro (+2) | 2 | Unrelated (+0) | 0 | Phylogenetically related (+2) | 2 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 64 | from Citrus sinensis, pH 7.5, 20°C, kinetic characterisation assay, | [10] |
Vmax values
| Vmax | Unit | Directionality | Organism | Organism | References |
|---|---|---|---|---|---|
| 0.08 | µmol/min/mg | forward | Cannabis sativa L. | [1] | |
| 0.12 ± 0.01 | µmol/min/mg | forward | Cannabis sativa | [1] | |
| 0.4748 | µmol/min/mg | forward | Citrus limon | References | |
| 0.53 | µM/min | forward | Citrus sinensis | 200µM GPP, 20°C, [E]: 0.07µM, pH7.5 | [10] |
| 0.64 | µM/min | forward | Citrus sinensis | 200µM GPP, 37°C, [E]: 0.26µM, pH7.5 | [10] |
| 19 | µmol/h/mg | forward | Mentha x piperita & Mentha spicata | Maximum specific activity, 1-10µg protein per mixture, pH &.0, 30°C | References |
Kcat values
| Parameter | Direction | Substrate | Value | Unit | Method | M_w | Organism | O_w | Expression_vector | E_w | Enzyme | En_w | pH | P_w | Temperature | T_w | Total_weight | Description | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kcat | Forward | GPP | 0.0004 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp, expressed in E.coli and truncated (R59), pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0004 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli , truncated R58P59, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0004 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli , truncated R58A59, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0004 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli , S60, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.002 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli preprotein, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0024 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli wt, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0034 | 1/s | in vitro (+2) | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli E57, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0036 | 1/s | in vitro | 2 | Unrelated (+0) | 0 | E.coli (+4) | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli Q54, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.0037 | 1/s | in vitro | 2 | Unrelated (+0) | 0 | E.coli | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli R58, pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.02 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Protein from Mentha sp. expressed in E.coli recombinant preprotein , pH7.0, 1-50 mg [E], 25 µM [GPP], limonene produced 94%, 2% myrcene, pinene | Williams1998 |
| Kcat | Forward | GPP | 0.04 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | close range (+2) | 2 | 37°C (+4) | 4 | 256 | Citrus sinensis, expressed in E.coli 200µM GPP, 37°C, [E]: 0.26µM, pH7.5 | Entova2013 |
| Kcat | Forward | GPP | 0.082 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | close range (+2) | 2 | 36°C - 40°C (+2) | 2 | 128 | Cannabis sativa L., expressed in E.coli,CsTPS1, pH6.5, 40°C, 1.25 mg [LimSynth] per 500ml assay mixture, 10mM [GPP] | Gunnewich2008 |
| Kcat | Forward | GPP | 0.09 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | close range (+2) | 2 | 36°C - 40°C (+2) | 2 | 128 | Cannabis sativa L., expressed in E.coli,Km of 6.25 ± 0.41 µM, pH 6.5, 40°C, [GPP] 0.5 - 25.0 µM used for kinetic parameter determination, kcat 0.09 /s | Gunnewich2008 |
| Kcat | Forward | GPP | 0.13 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | close range (+2) | 2 | 36°C - 40°C (+2) | 2 | 128 | Cannabis sativa L., expressed in E.coli,Km of 6.25 ± 0.41 µM, pH 6.5, 40°C, [GPP] 0.5 - 25.0 µM used for kinetic parameter determination, kcat 0.09 /s | Gunnewich2008 |
| Kcat | Forward | GPP | 0.13 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | close range (+2) | 2 | different temperature (+0) | 0 | 128 | Citrus sinensis, expressed in E.coli 200µM GPP, 20°C, [E]: 0.07 mM, pH7.5 | Entova2013 |
| Kcat | Forward | GPP | 0.3 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | different temperature (+0) | 0 | 128 | Mentha piperita & Mentha spicata, expressed in E.coli , pH7.0, 30°C, enzyme assay, 1 -10 mg protein per mixture | Alonso1992 |
| Kcat | Forward | GPP | 0.000039 | 1/s | in vitro | 2 | Unrelated | 0 | E.coli | 4 | Identical (+4) | 4 | 7.0 (+4) | 4 | 37°C (+4) | 4 | 512 | Citrus sinensis, expressed in E.coli40mM GPP, 37°C, [E]: 0.3 mM, pH7.0 (check units) | Olsen2011 |
Extracting Information from Limonene Production Rates
The production rates would reflect on the flux for this enzyme, and this would provide provide the insights on the Vmax of this enzyme.
| Amount produced (mg/L) | Time (H) | Organism | Description | Reaction Flux (µM/s) |
|---|---|---|---|---|
| 5 | 24 | Escherichia coli | Possible reason for the low limonene production might due to the insufficient supply of IPP and DMAPP [12]. | 0.0255 |
| 335 | 48 | Escherichia coli | Engineered E.coli in which heterologous MVA pathway was installed [13]. | 0.8537 |
| 35.8 | 48 | Escherichia coli | E.coli was engineered to express GPPS, LS, DXS, and IDI [14] . | 0.0912 |
| 4.87 | 48 | Escherichia coli | This was the initial titer. The study established a limonene biosynthesis pathway in E.coli using four different polycistronic operons based on 3 vectors with varied expression strength [14]. | 0.0124 |
| 17.4 | 48 | Escherichia coli | Using a plasmid with DXS and IDI over expressed [14]. | 0.0445 |
| 430 | 72 | Escherichia coli | [13] | 0.7306 |
Parameter estimation
This section can be found HERE
Simulations
Simulations performed can be found HERE.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Gunnewich, N. 2008. "Expression and characterization of terpene synthases from Cannabis sativa L. and Salvia sclarea L. Doctoral thesis. Cite error: Invalid
<ref>tag; name "Gunnewich2008" defined multiple times with different content - ↑ Turner,G. et. al.1999. "Limonene synthase, the enzyme responsible for monoterpene biosynthesis in peppermint, is localized to leucoplasts of oil gland secretory cells", Plant Physiology 120(3): 879-886
- ↑ Maruyama, T. et. al. 2002. "Molecular cloning, functional expression and characterization of d-Limonene synthase from Agastache rugosa" Biol. Pharm. Bull. 25(5): 661-665
- ↑ 4.0 4.1 4.2 Alonso et. al. 1992. "Purification of 4S-Limonene Synthase, a Monoterpene Cyclase from the Glandular Trichomes of Peppermint (Mentha x piperita) and Spearmint (Mentha spicata)", The Journal of Biological Chemistry, 267(11):7582-7587
- ↑ Latendresse M. (2013). "Computing Gibbs Free Energy of Compounds and Reactions in MetaCyc."
- ↑ 6.0 6.1 6.2 Olsen, S.N. 2011. "Isolation, Purification, and Characterization of (+)-4R-limonene synthase: The first step in exploring enzyme stereospecificity in terpenoid biosynthesis",Masters thesis, Brandeis university Cite error: Invalid
<ref>tag; name "Olsen2011" defined multiple times with different content Cite error: Invalid<ref>tag; name "Olsen2011" defined multiple times with different content - ↑ 7.00 7.01 7.02 7.03 7.04 7.05 7.06 7.07 7.08 7.09 7.10 7.11 Williams, D.C. et. al. 1998. "Truncation of Limonene Synthase preprotein provides a fully active 'pseudomature' form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair ",Biochemistry, 37:12213-12220 Cite error: Invalid
<ref>tag; name "Williams1998" defined multiple times with different content - ↑ Gunnewich, N., et al. 2006."Functional expression and characterization of trichome-specific (-)-limonene synthase and (+)-α-pinene synthase from Cannabis sativa", Natural Product Communications, 0(0): pp1-10
- ↑ 9.0 9.1 9.2 9.3 Lücker, J. et. al. 2002."Monoterpene biosynthesis in lemon (Citrus limon). cDNA isolation and functional analysis of four monoterpene synthases", Eur. J. Biochem. 269: pp3160-3171
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 Entova, S. 2013. "Kinetic characterization, crystallization, and photosynthetic expression of (+)-4R-limonene synthase from C. sinensis", Masters Thesis, Brandeis Univeristy Cite error: Invalid
<ref>tag; name "Entova2013" defined multiple times with different content - ↑ Adam, K. et. al. 1996. "Partial purification and characterization of a monoterpene cyclase, limonene synthase, from the liverwort Ricciocarpos natans. 332(2):pp 352-356.
- ↑ Carter, Ora A. et. al.2013. "Monoterpene biosynthesis pathway construction in Escherichia coli",Phytochemistry, 64:425–433, 2003.
- ↑ 13.0 13.1 Alonso-Gutierez et. al. 2013. "Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production", Metabolic Engineering, 19:33-41 Cite error: Invalid
<ref>tag; name "AlonsoGutierez2013" defined multiple times with different content - ↑ 14.0 14.1 14.2 Du et. al. 2014. "Enhanced limonene production by optimizing the expression of limonene biosynthesis and MEP pathway genes in E.coli", Bioprocessing and Bioprocessing, 1:10
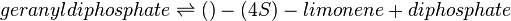
![V_\mathrm{LimSynth} = Vmax_\mathrm{forward} * \cfrac {\cfrac{[GPP]}{Km_\mathrm{GPP}} * \left ( 1 - \cfrac {[Limonene]*[PP]}{[GPP]*K_\mathrm{eq}} \right )}{1 + \cfrac {[GPP]}{Km_\mathrm{GPP}} + \cfrac {[Limonene]}{Km_\mathrm{Limonene}} + \cfrac {[PP]}{Km_\mathrm{PP}} + \cfrac {[Limonene]*[PP]}{Km_\mathrm{Limonene}*Km_\mathrm{PP}}}](/wiki/images/math/6/9/8/698f16353b522504a844aae5c37bdac9.png)