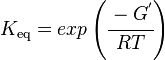Difference between revisions of "Limonene Synthase"
Aliah.hawari (talk | contribs) (→Kinetic Parameter Values) |
Aliah.hawari (talk | contribs) (→Kinetic Parameter Values) |
||
| Line 160: | Line 160: | ||
| align="center" style="background:#f0f0f0;"|'''References''' | | align="center" style="background:#f0f0f0;"|'''References''' | ||
|- | |- | ||
| − | | Km_gpp_LIMS||47.4||3.8||128||0||LIMS gene from Lavandula angustifolia was expressed in E. coli. The kinetics were measured in vitro at 30°C||Landmann2007 | + | | Km_gpp_LIMS||47.4||3.8||128||0||LIMS gene from Lavandula angustifolia was expressed in E. coli. The kinetics were measured in vitro at 30°C||<ref name="Landmann2007">[https://www.sciencedirect.com/science/article/pii/S0003986107003013 Landmann, C., et al.] (2007). "Cloning and characterization of three terpene synthases from lavender (''Lavandula angustifolia'')." Archives of Biochemistry and Biophysics 465: 417-429.</ref> |
|- | |- | ||
| Km_gpp_LIMS||130||NaN||32||0||LIMS gene from Citrus sinensis (orange) was expressed in E. coli. The Km was measured in vitro at 20°C.||Entova2013 | | Km_gpp_LIMS||130||NaN||32||0||LIMS gene from Citrus sinensis (orange) was expressed in E. coli. The Km was measured in vitro at 20°C.||Entova2013 | ||
Revision as of 12:51, 29 November 2018
You can go back to main page of the kinetic model here.
In this model, LIMS is modelled in Escherichia coli and this model is replicating the bacterial system in vivo. As such, in vivo-like conditions such as pH of 7.5 and temperature of 30°C in E. coli is set as the ideal conditions when assigning weights to its parameter. Limonene synthase catalyses the formation of limonene and pyrophosphate from one molecule of geranyl diphosphate (GPP). The following equations show LIMS’s reaction stoichiometry and its corresponding reaction rate using the Michaelis-Menten rate law:
Contents
Equation Rate
The reaction rate for LIMS is modelled using the reversible Michaelis-Menten equation, and is shown below:
where :
| Parameter | Description | Units |
|---|---|---|
| VLimSynth | Reaction rate for Limonene Synthase | μM/min |
| KcatLIMS | Turnover number for limonene synthase | min-1 |
| KmGPP | Michaelis-Menten constant for GPP | μM |
| KmLimonene | Michaelis-Menten constant for Limonene | μM |
| KmPP | Michaelis-Menten constant for PP | μM |
| Keq | Equilibrium constant | dimensionless |
| [GPP] | GPP concentration | μM |
| [Limonene] | Limonene concentration | μM |
| [PP] | PP concentration | μM |
Metabolite Background Information
Long metabolite names are abbreviated in the model for clarity and standard identification purposes.
| Metabolite | Abbreviation | Chemical Formula | Molar mass (g/mol) | ChEBI | ChEMBL | PubChem |
|---|---|---|---|---|---|---|
| geranyl diphosphate | GPP | C10H20O7P2 | 314.209 | 17211 | 41432 | 445995 |
| (-)-4S-limonene | Limonene | C10H16 | 136.24 | 15384 | 449062 | 22311 or 439250 |
| diphosphate | PP | O7P2 | 173.94 | 644102 | ||
| limonene synthase | LimSynth | 70.03 kDa [1], 72.4 kDa [2] ; 60kDa [3]; 56 kDa [4] |
Parameterisation
Calculating the Equilibrium Constant
Unlike the kinetic parameter values, thermodynamic parameter values such as for equilibrium constant (Keq) are not easily found in literature reports. However, Keq can be calculated from Gibbs Free Energy (ΔG°) using the following equation:
where;
| Keq | Equilibrium constant |
| -ΔG° | Gibbs free energy change (kcal/mol) |
| R | Gas constant (0.0019859 kcal/K/mol) |
| T | Absolute temperature (298 K) |
Gibbs free energy values for LIMS are obtained from MetaCyc (EC 4.2.3.16) is -28.049988 kcal/mol [5] and Equilibrator [1]. Table 2 summarizes the ΔrG° values found for LIMS and the calculated Keq. These Keq values are given an arbitrary equal weight of 1 as the ΔrG° values obtained were calculated from a group contribution method and do not have any measurement conditions that would allow us to assess the Keq values according to the weighting scheme set out in (insert link here).
| ΔrG°(kcal/mol) | Keq | Error (±) | Source | Weight |
|---|---|---|---|---|
| -28.050 | 3.843E+20 | N/A | MetaCyc | 1 |
| -42.161 ± 2.844 | 8.711E+30 | 5.876E+29 | Equilibrator | 1 |
Kinetic Parameter Values
| Parameter | Value | Error | Weight | Error type | Description | References |
| Km_gpp_LIMS | 47.4 | 3.8 | 128 | 0 | LIMS gene from Lavandula angustifolia was expressed in E. coli. The kinetics were measured in vitro at 30°C | [6] |
| Km_gpp_LIMS | 130 | NaN | 32 | 0 | LIMS gene from Citrus sinensis (orange) was expressed in E. coli. The Km was measured in vitro at 20°C. | Entova2013 |
| Km_gpp_LIMS | 6.8 | NaN | 32 | 0 | LIMS gene from Cannabis sativa L. var. 'Skunk' plants was expressed in E. coli. The kinetics were measured at 40°C. | [7] |
| Km_gpp_LIMS | 0.7 | NaN | 128 | 0 | LIMS gene from Citrus limon (lemon) was expressed in E. col. Kinetics were measured in vitro at 30°C. | Lucker2002 |
| Km_gpp_LIMS | 1.25 | NaN | 16 | 0 | LIMS isolated from Ricciocarpos natans. Kinetics measured at 32°C and pH7.0. | [8] |
| Km_gpp_LIMS | 1.8 | NaN | 32 | 0 | LIMS isolated from Mentha x piperita (peppermint). Kinetics were measured at 30°C. | Rajaonarivony1992 |
BRENDA data
To further enrich the kinetic parameter values for LIMS, parameter values from EC 4 to EC 4.2.3.* that can be obtained from BRENDA is downloaded. These BRENDA data is integrated with the rest of the kinetic parameter values using our ‘BRENDA Add-on’ protocol. In the BRENDA Add-On protocol, we’ve specified six different ‘EC cases’ that are arranged in order of rank. These EC cases are essentially six different datasets of parameter values downloaded from BRENDA that are filtered according to the specific enzyme class and organism of interest. For this case study example, six different ‘EC case’ datasets were downloaded from BRENDA each for Km and Kcat parameters (Tables below).
| Parameter value | Uncertainty* | Weight | Uncertainty type** |
|---|---|---|---|
| 6.80 | 14.9566 | 11.9024 | 1 |
| 49 | 69.0555 | 7.4390 | 1 |
| 121 | 36.1126 | 2.9756 | 1 |
| 500 | 36.9586 | 1.4878 | 1 |
| 300 | 23.8770 | 0.5951 | 1 |
| Parameter value | Uncertainty* | Weight | Uncertainty type** |
|---|---|---|---|
| 2.040 | 38.2439 | 9.5610 | 1 |
| 17.400 | 109.1960 | 5.9756 | 1 |
| 200.698 | 177.0428 | 2.3902 | 1 |
| 192.000 | 44.6715 | 1.1951 | 1 |
| 168.000 | 62.4126 | 0.4780 | 1 |
Parameter estimation
This section can be found HERE
Simulations
Simulations performed can be found HERE.
References
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedGunnewich2008 - ↑ Turner,G. et. al.1999. "Limonene synthase, the enzyme responsible for monoterpene biosynthesis in peppermint, is localized to leucoplasts of oil gland secretory cells", Plant Physiology 120(3): 879-886
- ↑ Maruyama, T. et. al. 2002. "Molecular cloning, functional expression and characterization of d-Limonene synthase from Agastache rugosa" Biol. Pharm. Bull. 25(5): 661-665
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedAlonso1992 - ↑ Latendresse M. (2013). "Computing Gibbs Free Energy of Compounds and Reactions in MetaCyc."
- ↑ Landmann, C., et al. (2007). "Cloning and characterization of three terpene synthases from lavender (Lavandula angustifolia)." Archives of Biochemistry and Biophysics 465: 417-429.
- ↑ Günnewich, N., Page, J.E., Köllner, T.G., Degenhardt, J., & Kutchan, T.M 2007. "Functional expression and characterization of trichome-specific (-)-limonene synthase and (+)-α-pinene synthase from Cannabis sativa ". Nat. Prod. Comm. 2(3): 223-232.
- ↑ Adam, K.-P., et al. (1996). "Partial purification and characterization of a monoterpene cyclase, limonene synthase, from the liverwort Ricciocarpus natans." Archives of Biochemistry and Biophysics 332(2): 352-356.
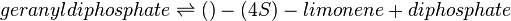
![V_\mathrm{LimSynth} = Kcat_\mathrm{LIMS}*[LIMS] * \cfrac {\cfrac{[GPP]}{Km_\mathrm{GPP}} * \left ( 1 - \cfrac {[Limonene]*[PP]}{[GPP]*K_\mathrm{eq}} \right )}{1 + \cfrac {[GPP]}{Km_\mathrm{GPP}} + \cfrac {[Limonene]}{Km_\mathrm{Limonene}} + \cfrac {[PP]}{Km_\mathrm{PP}} + \cfrac {[Limonene]*[PP]}{Km_\mathrm{Limonene}*Km_\mathrm{PP}}}](/wiki/images/math/b/c/3/bc34cd8a82726466dee4043dc9cc6bf8.png)