Difference between revisions of "Lactate dehydrogenase"
| Line 8: | Line 8: | ||
==Rate equation== | ==Rate equation== | ||
RAndom Bi-Bi reversible Michaelis-Menten equation is used. <ref name="Hernandez2011"> Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 ([http://dx.doi.org/10.1016/j.bbabio.2010.11.006 doi]) </ref> | RAndom Bi-Bi reversible Michaelis-Menten equation is used. <ref name="Hernandez2011"> Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 ([http://dx.doi.org/10.1016/j.bbabio.2010.11.006 doi]) </ref> | ||
| + | |||
<center><math> \frac{V_{mf}\frac{[NADH][PYR]}{Km_{NADH} K_{PYR}} - V_{mr}\frac{[Lactate_{in}][NAD]}{Km_{Lactate_{in}} K_{NAD}}}{1 + \frac{[NADH]}{Km_{NADH}} + \frac{[PYR]}{Km_{PYR}} + \frac{[NADH][PYR]}{Km_{NADH} Km_{PYR}} + \frac{[Lactate_{in}][NAD]}{Km_{Lactate_{in}} Km_{NAD}} + \frac{[Lactate_{in}]}{Km_{Lactate_{in}}} + \frac{[PYR]}{Km_{PYR}} } </math></center> | <center><math> \frac{V_{mf}\frac{[NADH][PYR]}{Km_{NADH} K_{PYR}} - V_{mr}\frac{[Lactate_{in}][NAD]}{Km_{Lactate_{in}} K_{NAD}}}{1 + \frac{[NADH]}{Km_{NADH}} + \frac{[PYR]}{Km_{PYR}} + \frac{[NADH][PYR]}{Km_{NADH} Km_{PYR}} + \frac{[Lactate_{in}][NAD]}{Km_{Lactate_{in}} Km_{NAD}} + \frac{[Lactate_{in}]}{Km_{Lactate_{in}}} + \frac{[PYR]}{Km_{PYR}} } </math></center> | ||
| + | |||
| + | Modified rate law to take Thermodynamic constraint into consideration | ||
| + | <center><math> \frac{V_{mf}\frac{[NADH][PYR]}{Km_{NADH} K_{PYR}} \left(1 - \frac{[Lactate_{in}][NAD]}{K_{eq}[NADH][PYR]} \right)}{1 + \frac{[NADH]}{Km_{NADH}} + \frac{[PYR]}{Km_{PYR}} + \frac{[NADH][PYR]}{Km_{NADH} Km_{PYR}} + \frac{[Lactate_{in}][NAD]}{Km_{Lactate_{in}} Km_{NAD}} + \frac{[Lactate_{in}]}{Km_{Lactate_{in}}} + \frac{[PYR]}{Km_{PYR}} } </math></center> | ||
==Prameter values== | ==Prameter values== | ||
Revision as of 11:31, 20 June 2014
A dehydrogenase is an enzyme that transfers a hydride from one molecule to another. Lactate dehydrogenase catalyzes the conversion of pyruvate to lactate and back, as it converts NADH to NAD+ and back.
Contents
Chemical reactions
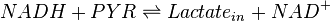
Rate equation
RAndom Bi-Bi reversible Michaelis-Menten equation is used. [1]
![\frac{V_{mf}\frac{[NADH][PYR]}{Km_{NADH} K_{PYR}} - V_{mr}\frac{[Lactate_{in}][NAD]}{Km_{Lactate_{in}} K_{NAD}}}{1 + \frac{[NADH]}{Km_{NADH}} + \frac{[PYR]}{Km_{PYR}} + \frac{[NADH][PYR]}{Km_{NADH} Km_{PYR}} + \frac{[Lactate_{in}][NAD]}{Km_{Lactate_{in}} Km_{NAD}} + \frac{[Lactate_{in}]}{Km_{Lactate_{in}}} + \frac{[PYR]}{Km_{PYR}} }](/wiki/images/math/4/b/2/4b2b29cf3850c4c61d11218132ecd486.png)
Modified rate law to take Thermodynamic constraint into consideration
![\frac{V_{mf}\frac{[NADH][PYR]}{Km_{NADH} K_{PYR}} \left(1 - \frac{[Lactate_{in}][NAD]}{K_{eq}[NADH][PYR]} \right)}{1 + \frac{[NADH]}{Km_{NADH}} + \frac{[PYR]}{Km_{PYR}} + \frac{[NADH][PYR]}{Km_{NADH} Km_{PYR}} + \frac{[Lactate_{in}][NAD]}{Km_{Lactate_{in}} Km_{NAD}} + \frac{[Lactate_{in}]}{Km_{Lactate_{in}}} + \frac{[PYR]}{Km_{PYR}} }](/wiki/images/math/e/5/4/e54419f6b7939faa1e16564482b31638.png)
Prameter values
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
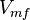
|
3.4 [1] | 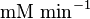
|
HeLa cell line | |
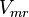
|
0.54 | 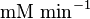
|
HeLa cell line | |
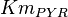
|
0.1 | mM | HeLa cell line | |
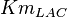
|
4.7 | mM | Rat AS-30D hepatoma | |
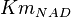
|
0.07 | mM | HeLa cell line | |
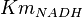
|
0.002 | mM | HeLa cell line |
Parameters with uncertainty
- Mean and Std. Dev. for
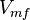 has been reported in Table S3 for Marín-Hernández (2011) et. al. [1]. The Std. Dev. for
has been reported in Table S3 for Marín-Hernández (2011) et. al. [1]. The Std. Dev. for 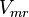 is calculated based on the same ratio for
is calculated based on the same ratio for 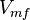 .
.
- The value for
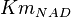 and
and 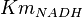 has been reported for Ovine (Sheep) as
has been reported for Ovine (Sheep) as 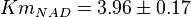 and
and 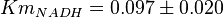 [4]. Due to lack of data in Human cells these two values are considered in our model.
[4]. Due to lack of data in Human cells these two values are considered in our model.
-
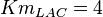 for LDH-1 and 2 isoforms and
for LDH-1 and 2 isoforms and 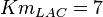 for LDH-4 and 5 isoforms are being reported in Marín-Hernández et. al. (2009)[5]. Mean and Std. Dev. from these two values are
for LDH-4 and 5 isoforms are being reported in Marín-Hernández et. al. (2009)[5]. Mean and Std. Dev. from these two values are 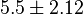
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
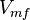
|
Failed to parse (Cannot store math image on filesystem.): 3.4 \pm 0.5 (3) [6] | 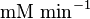
|
HeLa cell line | |
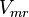
|
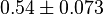
|
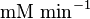
|
HeLa cell line | |
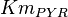
|
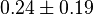
|
mM | HeLa cell line | |
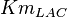
|
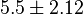
|
mM | Rat AS-30D hepatoma | |
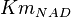
|
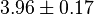
|
mM | Ovine (Sheep) | |
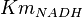
|
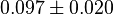
|
mM | Ovine (Sheep) |
References
- ↑ 1.0 1.1 1.2 1.3 Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 (doi)
- ↑ LeVan K.M., Goldberg E. (1991), Properties of human testis-specific lactate dehydrogenase expressed from Escherichia coli, Biochem. J. 273, 587-592 (1991)
- ↑ Pettit S.M., Nealon D.A., Henderson A.R. (1981), Purification of lactate dehydrogenase isoenzyme-5 from human liver, Clin. Chem. 27, 88-93 (1981)
- ↑ M. Doughty (1998), Some kinetic properties of lactate dehydrogenase activity in cell extracts from a mammalian (ovine) corneal epithelium, Exp. Eye Res., 66, pp. 231–239
- ↑ A. Marín-Hernández, J.C. Gallardo-Pérez, S.J. Ralph, S. Rodríguez-Enríquez, R. Moreno-Sánchez (2009), HIF-1alpha modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms, Mini Rev. Med. Chem., 9, pp. 1084–1101
- ↑ Marín-Hernández A , Rodríguez-Enríquez S, Vital-González P A, et al. (2006). Determining and understanding the control of glycolysis in fast-growth tumor cells. Flux control by an over-expressed but strongly product-inhibited hexokinase. FEBS J., 273 , pp. 1975–1988(doi)