Difference between revisions of "Hydroxymethyglutaryl-CoA synthase (HMGS)"
Aliah.hawari (talk | contribs) (→Keq for HMGS) |
Aliah.hawari (talk | contribs) (→Forward Kcat for HMGS) |
||
| Line 211: | Line 211: | ||
| style="width: 20%;" |[[File:Nagegowda_2004_HMGS.png|450px|Km values for Acetyl-CoA and Acetoacetyl-CoA]] | | style="width: 20%;" |[[File:Nagegowda_2004_HMGS.png|450px|Km values for Acetyl-CoA and Acetoacetyl-CoA]] | ||
| style="width: 10%;" |Gene from ''Brassica juncea'' and expressed in ''E. coli''. pH 8.5, 35"C. | | style="width: 10%;" |Gene from ''Brassica juncea'' and expressed in ''E. coli''. pH 8.5, 35"C. | ||
| − | | style="width: 5%;" |<ref name=Nagegowda2004 /> | + | | style="width: 5%;" |<ref name=Nagegowda2004/> |
|- | |- | ||
| 276 | | 276 | ||
Revision as of 14:44, 14 June 2019
You can go back to main page of the kinetic model here.
Hydroxymethyl-glutaryl synthase (HMGS, EC 2.3.3.10 ) is not native to E. coli and a heterologous HMGS gene from S. cerevisiae was engineered into the cell [1].
Contents
HMGS Parameter Values
HMGS is modelled using Bi-Bi Michaelis-Menten rate law as shown below:
Km for Acetyl-CoA
| Value | SD or SEM | Unit | Weight | Where did I get that? | Notes | Reference |
|---|---|---|---|---|---|---|
| 43 | NaN | μM | 8 | 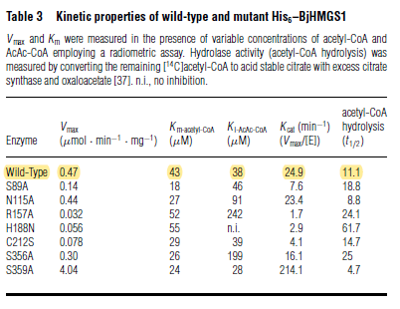
|
Gene from Brassica juncea (indian mustard) and expressed in E. coli. pH 8.5, 35°C. | [2] |
| 14 | NaN | μM | 64 | 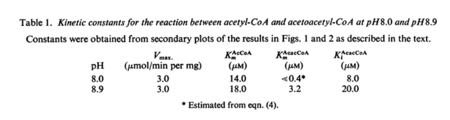
|
pH 8.0; 30°C, gene from Saccharomyces cerevisiae | [3] |
| 18 | NaN | μM | 64 | 
|
pH 8.9; 30°C, gene from Saccharomyces cerevisiae | [3] |
| 15.3 | NaN | μM | 64 | 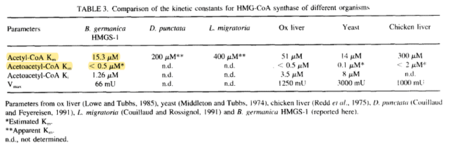
|
pH 8.5, 30C, 2 cytosolic HMG-CoA synthase genes from Blatella germanica (cockroach), HMGS-1 and HMGS-2 described, HMGS-1 cloned in expression vector pSBLA1 and overexpressed in Escherichia coli BL21DE3 pLys S; intronless gene, encodes for an active enzyme able to complement , eukaryotic Mev-1 cells. Measured at 30°C, initiated by adding 0.2-0.6 microgram of protein, final volume 400 microlitre, pH 7.0- 8.5. | [4] |
| 50 | μM | 16 | 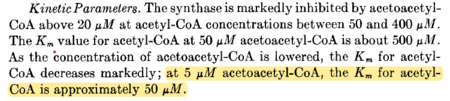
|
from chicken liver, pH 8.2, 30C, measured using spectrophometric assay. | [5] | |
| 50 | 6 | μM | 16 | 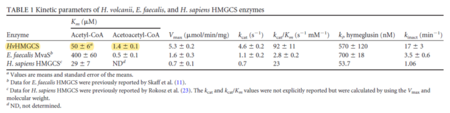
|
Gene from H. volcanii, expressed in H. volcanii. pH 8.0, 30C. Uncertainty measured as SEM. | [6] |
| 270 | NaN | μM | 16 | 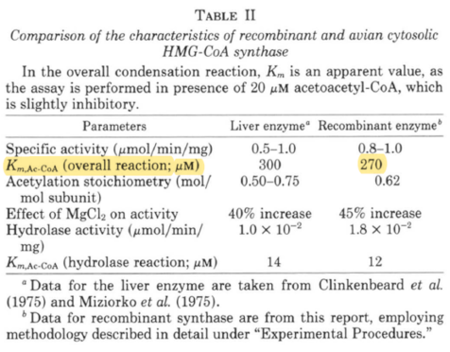
|
Gene from Avian liver, expressed in E. coli, pH 8.2, 30°C | [7] |
| 290 | 22 | μM | 16 | 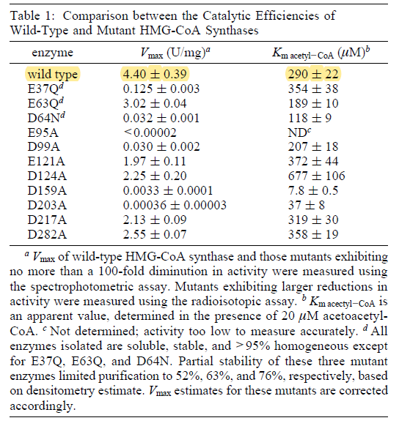
|
gene from E. coli, expressed in E. coli, pH 8.2, 30C. Uncertainty measured as SE. | [8] |
| 294 | NaN | μM | 16 | 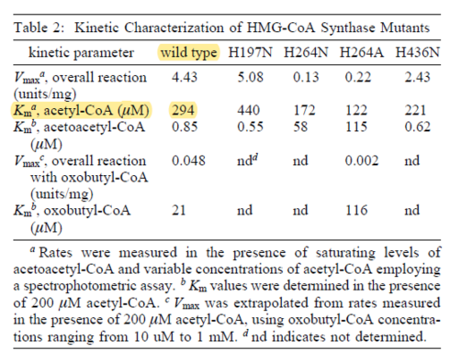
|
avian enzyme;wildtype and mutants; expression vector pET-3d, cloned and expressed in Escherichia coli BL21 (DE3), pH 8.2, temperature not mentioned. | [9] |
| 350 | NaN | μM | 4 | 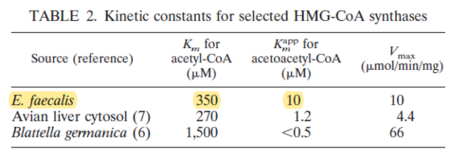
|
Gene from E. faecalis, expressed in E. coli, pH 9.8, 37C. | [10] |
Km for Acetoacetyl-CoA
| Value | SD or SEM | Unit | Weight | Where did I get that? | Notes | Reference |
|---|---|---|---|---|---|---|
| 0.35 | NaN | μM | 4 | 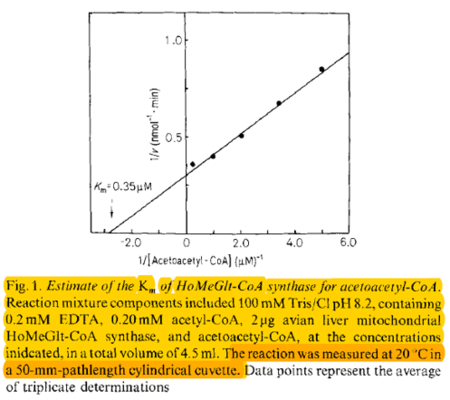
|
Gene from Gallus gallus, assay pH 8.2, 20°C. | [11] |
| 0.4 | NaN | μM | 64 | 
|
pH 8.0; 30°C, gene from Saccharomyces cerevisiae, kinetics measured at different pHs. Uncertainty measurement not mentioned. | [3] |
| 3.2 | NaN | μM | 64 | 
|
pH 8.9; 30°C, gene from Saccharomyces cerevisiae, kinetics measured at different pHs. Uncertainty measurement not mentioned. | [3] |
| 0.5 | NaN | μM | 16 | 
|
pH 8.5, 30C, 2 cytosolic HMG-CoA synthase genes from Blatella germanica, HMGS-1 and HMGs-2 described, HMGS-1 cloned in expression vector pSBLA1 and overexpressed in Escherichia coli BL21DE3 pLys S; intronless gene, encodes for an active enzyme able to complement , eukaryotic Mev-1 cells. Measured at 30°C, initiated by adding 0.2-0.6 microgram of protein, final volume 400 microlitre, pH 7.0- 8.5. | [4] |
| 0.85 | naN | μM | 4 | 
|
Avian enzyme. wildtype and mutants; expression vector pET-3d, cloned and expressed in Escherichia coli BL21 (DE3), pH 8.2, temperature not mentioned. | [9] |
| 1.19 | 0.12 | μM | 16 | 
|
Gene from E. coli, expressed in E. coli, pH 8.2, 30°C | [8] |
| 1.4 | 0.2 | μM | 16 | 
|
Gene from H. volcanii, expressed in H. volcanii. pH 8.0, 30°C | [6] |
| 10 | NaN | μM | 4 | 
|
Gene from E. faecalis, expressed in E. coli, pH 9.8, 37C. | [10] |
Forward Kcat for HMGS
| Value | SD or SEM | Weight | Unit | Where did I get that? | Notes | References |
|---|---|---|---|---|---|---|
| 24.9 | NaN | 8 | 1/min | Km values for Acetyl-CoA and Acetoacetyl-CoA | Gene from Brassica juncea and expressed in E. coli. pH 8.5, 35"C. | [12] |
| 276 | 12 | 16 | 1/min | 
|
Gene from H. volcanii, expressed in H. volcanii. pH 8.0, 30C. Uncertainty measured as SEM. | [6] |
Keq for HMGS
| Value | ΔG°' (kcal/mol) | Weight | Where did I get that? | Reference |
|---|---|---|---|---|
| 1.08E+07 | -9.58 | 1 | [13] | |
| 9.17E+02 | -4.04 | 5 | [14] |
HMGS parameter distribution properties
References
Template:Reflist- ↑ Alonso-Gutierrez, J., et al.(2013). "Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production." Metabolic Engineering 19: 33-41.
- ↑ Nagegowda, D. A., et al. (2004). "Brassica juncea 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase 1: expression and characterization of recombinant wild-type and mutant enzymes." Biochem. J. 383: 517-527.
- ↑ 3.0 3.1 3.2 3.3 Middleton, B. (1972). "The kinetic mechanism of 3-Hydroxy-3-methylglutaryl-Coenzyme A Synthase from baker's yeast." Biochem. J. 126: 35-47.
- ↑ 4.0 4.1 Cabano, J., et al. (1997). "Catalytic properties of recombinant 3-Hydroxy-3-methylglutaryl Coenzyme A synthase-1 from Blatella Germanica." Insect Biochem. Molec. Biol. 27(6): 499-505.
- ↑ Reed, W. D. and M. D. Lane (1975). "Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase from chicken liver." Methods in Enzymology 35: 155-160.
- ↑ 6.0 6.1 6.2 VanNice, J. C., et al. (2013). "Expression in Haloferax volcanii of 3-Hydroxymethylglutaryl Coenzyme A synthase facilitates isolation and characterization of the active form of a key enzyme required for polyisoprenoid cell membrane biosynthesis in halophilic archaea." Journal of Bacteriology 195(17): 3854-3862.
- ↑ Misra, I., et al. (1993). "Avian 3-hydroxy-3-methylglutaryl-CoA synthase." The Journal of Biological Chemistry 288(16): 12129-12135.
- ↑ 8.0 8.1 Chun, K. Y., et al. (2000). "3-hydroxy-3-methylglutaryk-CoA synthase: participation of invariant acidic residues in formation of the acetyl-S-enzyme reaction intermediate." Biochemistry 39: 14670-14681.
- ↑ 9.0 9.1 Misra, I. and H. M. Miziorko (1996). "Evidence for the interaction of avian 3-hydroxy-3-methylglutaryl-CoA synthase histidine 264 with acetoacetyl-CoA." Biochemistry 35: 9610-9616.
- ↑ 10.0 10.1 Sutherlin, A., et al. (2002). "Enterococcus faecalis 3-Hydroxy-3-methylglutaryl coenzyme A synthase, an enzyme of isopentenyl diphosphate biosynthesis." Journal of Bacteriology 184(15): 4065-4070.
- ↑ Menahan, L. A., et al. (1981). "Interrelationships between 3-Hydroxy-3-Methylglutaryl-CoA synthase, acetoacetyl-COA and ketogenesis." Eur. J. Biochem. 119: 287-294.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedNagegowda2004 - ↑ Latendresse, M. (2013). "Computing Gibbs free energy of compounds and reactions in MetaCyc." from https://biocyc.org/PGDBConceptsGuide.shtml#gibbs.
- ↑ E. Noor, H.S. Haraldsdóttir, R. Milo, R.M.T. Fleming (2013) Consistent Estimation of Gibbs Energy Using Component Contributions PLoS Comput Biol 9(7): e1003098 [DOI: 10.1371/journal.pcbi.1003098]
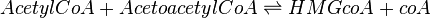
![V_\mathrm{HMGS} = Kcat_\mathrm{forward} * [HMGS] * \cfrac {\left ( \cfrac{[AcetylcoA]}{Km_\mathrm{AcetylcoA}} * \cfrac {[AcetoacetylcoA]}{Km_\mathrm{AcetoacetylcoA}} \right ) * \left ( 1 - \cfrac {[HMGcoA]*[coA]}{[AcetylcoA]*[AcetoacetylcoA]*K_\mathrm{eq}} \right )}
{ \left (1 + \cfrac {[AcetylcoA]}{Km_\mathrm{AcetylcoA}} + \cfrac {[coA]}{Km_\mathrm{coA}} \right ) + \left ( 1+ \cfrac {[AcetoacetylcoA]}{Km_\mathrm{AcetoacetylcoA}} + \cfrac {[HMGcoA]}{Km_\mathrm{HMGcoA}} \right ) }](/wiki/images/math/3/b/6/3b65b5bd28406dd4553b4b8fcd1cb152.png)