Difference between revisions of "Glycine out"
| Line 1: | Line 1: | ||
| − | [[Category: | + | [[Category:Uncertainty]] |
| − | |||
This reaction describes the utilization of the endproduct Glycine in other pathways. | This reaction describes the utilization of the endproduct Glycine in other pathways. | ||
| Line 12: | Line 11: | ||
==Parameters== | ==Parameters== | ||
| + | *For Transport reactions it is presumed to be at equilibrium, such that <math>K_{eq} = 1</math> and <math>K_{1} = K_{2}</math>. | ||
{|class="wikitable" | {|class="wikitable" | ||
! Parameter | ! Parameter | ||
| Line 23: | Line 23: | ||
|<math>S^{-1}</math> | |<math>S^{-1}</math> | ||
|Escherichia coli | |Escherichia coli | ||
| + | |- | ||
| + | |<math>K_{2}</math> | ||
| + | |<math>8.03 \times 10^{-3}</math> | ||
| + | |<math>S^{-1}</math> | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | ==Parameters with uncertainty== | ||
| + | *The transport rates have been modelled using mass action kinetics (i.e., as non-saturable, non-enzymatic reactions). No information is available about the uncertainty of these parameters. As these parameters are strictly positive, they are sampled using a log-normal distribution as are and values. The means of <math>K_{1}</math> are set to the value reported in Turnaev (2006) et al. <ref name="Turnaev_2006"></ref> for the fixed-parameter model. The reaction is presumed to be at equilibrium, such that <math>K_{eq} = 1</math> and <math>K_{1} = K_{2}</math>. The sampling of the parameters are done in a way so that it ranges between <math>[0.001\times mean \quad 1000 \times mean ]</math> to allow a large exploration of the parameter space. | ||
| + | {|class="wikitable" | ||
| + | ! Parameter | ||
| + | ! Value | ||
| + | ! Organism | ||
| + | ! Remarks | ||
| + | |- | ||
| + | |<math>K_{1}</math> | ||
| + | |Sampled between 0.00000803 and 8.03 | ||
| + | |rowspan="2"| | ||
| + | |rowspan="2"| | ||
| + | |- | ||
| + | |<math>K_{2}</math> | ||
| + | |Sampled between 0.00000803 and 8.03 | ||
|} | |} | ||
| + | |||
==References== | ==References== | ||
<references/> | <references/> | ||
Revision as of 11:49, 15 May 2014
This reaction describes the utilization of the endproduct Glycine in other pathways.
Contents
Reaction equation
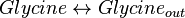
Rate equation
Simple mass action rate law is used.
![v = K_{1} * [Glycine] - K_{2} * [Glycine_{out}]](/wiki/images/math/6/d/4/6d41306edc0998827f9c39069eb7767a.png)
Parameters
- For Transport reactions it is presumed to be at equilibrium, such that
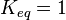 and
and 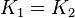 .
.
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
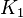
|
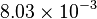 [1] [1]
|
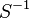
|
Escherichia coli | |
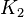
|
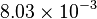
|
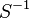
|
Parameters with uncertainty
- The transport rates have been modelled using mass action kinetics (i.e., as non-saturable, non-enzymatic reactions). No information is available about the uncertainty of these parameters. As these parameters are strictly positive, they are sampled using a log-normal distribution as are and values. The means of
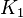 are set to the value reported in Turnaev (2006) et al. [1] for the fixed-parameter model. The reaction is presumed to be at equilibrium, such that
are set to the value reported in Turnaev (2006) et al. [1] for the fixed-parameter model. The reaction is presumed to be at equilibrium, such that 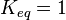 and
and 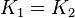 . The sampling of the parameters are done in a way so that it ranges between
. The sampling of the parameters are done in a way so that it ranges between ![[0.001\times mean \quad 1000 \times mean ]](/wiki/images/math/7/0/5/70565b13097c64cb12ee455eb2006ee7.png) to allow a large exploration of the parameter space.
to allow a large exploration of the parameter space.
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
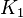
|
Sampled between 0.00000803 and 8.03 | ||
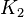
|
Sampled between 0.00000803 and 8.03 |