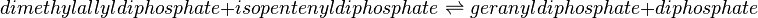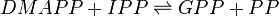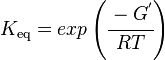Geranyl diphosphate synthase (GPPS)
You can go back to main page of the kinetic model here.
Model construction progress :
| Enzyme | Pathway | Keq | Kcat | VmaxF | KmIPP | KmDMAPP | [Sub1] | [Sub2] | VmaxR | KmGPP | KmPP | [Prod1] | [Prod2] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01_GPPS | Limonene Biosynthesis | 1 | 3 | 4 | 4 | 4 | 3 | 3 |
Legend:
| Have not started ·· | 1 -2 data found ·· | 3-4 data found ·· | sufficient data found/estimated ·· | data distribution generated ·· | data sampled |
| to do | DONE! |
Contents
What we know
Issues
Strategies
Reaction catalysed
Metabolite and Enzyme Background Information
Long metabolite names are abbreviated in the model for clarity and standard identification purposes.
| Metabolite | Abbreviation | Chemical Formula | Molar mass (g/mol) | ChEBI | ChEMBL | PubChem | MetaCyc | BRENDA |
|---|---|---|---|---|---|---|---|---|
| dimethylallyl diphosphate | DMAPP | |||||||
| isopentenyl diphosphate | IPP | |||||||
| geranyl diphosphate | GPP | C10H20O7P2 | 314.209 | 17211 | 41432 | 445995 | ||
| diphosphate | PP | O7P2 | 173.94 | 644102 | ||||
| geranyl diphosphate synthase | GPPS | 32.16 kD (from nucleotide sequence), 36 kD (experimental) | CPLX-8656 | 2.5.1.1 |
Equation Rate
- Failed to parse (Cannot store math image on filesystem.): V_\mathrm{GPPS} = Vmax_\mathrm{forward} * \cfrac { \left (\cfrac{[DMAPP]}{Km_\mathrm{DMAPP}} * \cfrac{[IPP]}{Km_\mathrm{IPP}}\right )* \left ( 1 - \cfrac {[GPP]*[PP]}{[GPP]*[PP]*K_\mathrm{eq}} \right )}{\left (1 + \cfrac {[IPP]}{Km_\mathrm{IPP}} + \cfrac {[PP]}{Km_\mathrm{PP}} \right ) * \left ( 1 + \cfrac {[DMAPP]}{Km_\mathrm{DMAPP}} + \cfrac {[GPP]}{Km_\mathrm{GPP}} \right )}
| Parameter | Description |
|---|---|
| VGPPS | Reaction rate for Geranyl diphosphate synthase |
| Vmaxforward | Maximum reaction rate towards the production of GPP |
| KmGPP | Michaelis-Menten constant for GPP |
| KmIPP | Michaelis-Menten constant for IPP |
| KmPP | Michaelis-Menten constant for PP |
| KmDMAPP | Michaelis-Menten constant for DMAPP |
| Keq | Equilibrium constant |
| [GPP] | GPP concentration |
| [DMAPP] | DMAPP concentration |
| [IPP] | DMAPP concentration |
| [PP] | PP concentration |
Strategies for estimating the kinetic parameter values
Calculating the Equilibrium Constant
Standard Gibbs Free energy for GPPS from MetaCyc is -13.0 kcal mol-1.
The equilibrium constant can be calculated using the Van't Hoff Isotherm equation:
Failed to parse (Cannot store math image on filesystem.): = exp \left ( \cfrac {-(-13.0 \text { kcalmol}^{-1})}{ (1.98722 \text{ calK}^{-1} \text { mol}^{-1} * 289 K} \right )
Failed to parse (Cannot store math image on filesystem.): = exp \left ( \cfrac {13.0 \text { kcalmol}^{-1} }{ 574.30658 \text{ calK}^{-1}\text { mol}^{-1} }\right)
Failed to parse (Cannot store math image on filesystem.): = exp \left ( \cfrac{ 13000 \text {calmol}^{-1}}{574.30658 \text{ cal}\text { mol}^{-1}} \right)
Failed to parse (Cannot store math image on filesystem.): = 6788526395
where;
| Keq | Equilibrium constant |
| -?G° | Gibbs free energy change. For GPPS it is -13.0 kcalmol-1 |
| R | Gas constant with a value of 1.98722 calK-1mol-1 |
| T | Temperature which is always expressed in kelvin, as a standard, I used 298 K |
Published Kinetic Parameter Values
Values for the kinetic parameter required to simulate this model can be obtained from published and unpublished literature.
A more detailed descriptions of the values listed above can be found HERE , where I've linked and highlighted where these data came from.
Substrate, Product & Enzyme Concentration Values
| Concentration | Unit | Substrate / Product | Directionality | Organism | References |
|---|---|---|---|---|---|
| 15 | µM | IPP | forward | Phaedon cochleariae | [1] |
| 50 | µM | DMAPP | forward | Phaedon cochleariae | [1] |
| 50 | µM | DMAPP | forward | Phaedon cochleariae | [1] |
| 50 | µM | IPP | forward | Phaedon cochleariae | [1] |
| 250 | µM | IPP | forward | Picea abies | [2] |
| 800 | µM | DMAPP | forward | Picea abies | [2] |
Km Values
| Km (mM) | Unit | Substrate / Product | Directionality | Organism | References |
|---|---|---|---|---|---|
| 81 | μM | DMAPP | forward | Mentha sp. | [3] |
| 88 | μM | IPP | forward | Mentha sp. | [3] |
| 241 | μM | DMAPP | forward | Mentha sp. | [3] |
| 215 | μM | DMAPP | forward | Mentha sp. | [3] |
| 44 | μM | IPP | forward | Abies grandis | [4] |
| 55 | μM | IPP | forward | Abies grandis | [4] |
| 44 | μM | IPP | forward | Abies grandis | [4] |
| 163 | μM | DMAPP | forward | Abies grandis | [4] |
| 90 | μM | DMAPP | forward | Abies grandis | [4] |
| 82 | μM | DMAPP | forward | Abies grandis | [4] |
| 0.84 | μM | IPP | forward | Phaedon cochleariae | [1] |
| 11.6 | μM | DMAPP | forward | Phaedon cochleariae | [1] |
| 1103 | μM | DMAPP | forward | Phaedon cochleariae | [1] |
| 11.79 | μM | IPP | forward | Phaedon cochleariae | [1] |
| 58 | μM | DMAPP | forward | Mycobacterium tuberculosis | [5] |
| 163 | μM | IPP | forward | Mycobacterium tuberculosis | [5] |
| 390 | μM | DMAPP | forward | Picea abies | [2] |
| 170 | μM | IPP | forward | Picea abies | [2] |
Vmax values
| Vmax | Unit | Directionality | Organism | References |
|---|---|---|---|---|
| 0.44 | µmol/min/mg (unit) | forward | Phaedon cochleariae | [1] |
| 0.67 | µmol/min/mg (unit) | forward | Phaedon cochleariae | [1] |
| 2.39 | µmol/min/mg (unit) | forward | Phaedon cochleariae | [1] |
| 0.17 | µmol/min/mg (unit) | forward | Phaedon cochleariae | [1] |
| 4.2 | nkat/mg | forward | Picea abies | [2] |
Kcat values
| Kcat | Unit | Organism | Reference |
|---|---|---|---|
| 0.7 | min-1 | Mycobacterium tuberculosis | [5] |
| 5.1 | min-1 | Mycobacterium tuberculosis | [5] |
| 1.4 | s-1 | Picea abies | [2] |
| 1.8 | s-1 | Abies grandis | [4] |
| 0.5 | s-1 | Abies grandis | [4] |
| 1.6 | s-1 | Abies grandis | [4] |
Extracting Information from (INSERT SUBSTRATE/PRODUCT) Production Rates
| Amount produced (mg/L) | Time (H) | Organism | Description | Reaction Flux (µM/s) |
|---|---|---|---|---|
| X | X | Y | Z | Z |
| X | X | Y | Z | Z |
| X | X | Y | Z | Z |
| X | X | Y | Z | Z |
| X | X | Y | Z | Z |
| X | X | Y | Z | Z |
Simulations
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 Frick, S. 2013. "Metal ions control product specificity of isoprenyl diphosphate synthases in the insect terpenoid pathway", PNAS, 110(11):4194-4199 Cite error: Invalid
<ref>tag; name "Frick2013" defined multiple times with different content Cite error: Invalid<ref>tag; name "Frick2013" defined multiple times with different content Cite error: Invalid<ref>tag; name "Frick2013" defined multiple times with different content Cite error: Invalid<ref>tag; name "Frick2013" defined multiple times with different content - ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Schmidt, A. 2010. "A bifunctional geranyl and geranylgeranyl diphosphate synthase is involved in terpene oleoresin formation in Picea abies", Plant Physiology, 152:639-655
- ↑ 3.0 3.1 3.2 3.3 Burke, C. 2004. "Heteromeric geranyl diphosphate synthase from mint: construction of a functional fusion protein and inhibition by biphosphonate substrate analogs", Archives of Biochemistry and Biophysics, 422:52-60
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 4.8 Burke, C. 2002. "Geranyl diphosphate synthase from Abies grandis:cDNA isolation, functional expression, and characterization", Archives of Biochemistry and Biophysics, 405:130-136
- ↑ 5.0 5.1 5.2 5.3 Mann, F.M. 2011. "Rv0989c encodes a novel (E)-geranyl diphosphate synthase facilitating decaprenyl diphosphate biosynthesis in Mycobacterium tuberculosis", FEBS Letters, 585(3):549-554