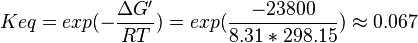Difference between revisions of "Fructose 1,6 bisphosphate aldolase"
| Line 47: | Line 47: | ||
==Parameters with uncertainty== | ==Parameters with uncertainty== | ||
| − | *The value for <math>V_{mf}</math> is collected from Hernandez ''et. al.'' <ref name="Hernandez_2006"></ref>. The <math>V_{mr}</math> is calcualted from the sampled <math>V_{mr}</math>, <math>Km_{Gly3P}</math>, <math>Km_{DHAP}</math> and <math>K_{eq}</math> values using the Haldane equation . | + | *The value for <math>V_{mf}</math> is collected from Hernandez ''et. al.'' <ref name="Hernandez_2006"></ref>. The <math>V_{mr}</math> is calcualted from the sampled <math>V_{mr}</math>, <math>Km_{Gly3P}</math>, <math>Km_{DHAP}</math> and <math>K_{eq}</math> values using the Haldane equation. '''Alternatively''' the reported value from AS-30D can be considered. The standard deviation can be calculated based on the same ratio of <math>V_{mf}</math>. However, as it is from a different species an extra 20% of uncertainty will be added. This gives the value <math>V_{mf}=0.063 \pm 0.028 </math> |
*The value for <math>Km_{Fru1,6BP}, Km_{Gly3P}, Km_{DHAP}</math> are collected from Ali D. Malay ''et. al.'' <ref name="Ali">Malay AD, Procious SL, Tolan DR. (2002). ''The temperature dependence of activity and structure for most prevalent mutant aldolase B associated with hereditary fructose intolerance'', Arch BiochemBiophys 408: 295–304.</ref> for wildtype Aldolase B gene at <math>30^{\circ}</math>C. | *The value for <math>Km_{Fru1,6BP}, Km_{Gly3P}, Km_{DHAP}</math> are collected from Ali D. Malay ''et. al.'' <ref name="Ali">Malay AD, Procious SL, Tolan DR. (2002). ''The temperature dependence of activity and structure for most prevalent mutant aldolase B associated with hereditary fructose intolerance'', Arch BiochemBiophys 408: 295–304.</ref> for wildtype Aldolase B gene at <math>30^{\circ}</math>C. | ||
| Line 64: | Line 64: | ||
|- | |- | ||
|<math>V_{mr}</math> | |<math>V_{mr}</math> | ||
| − | |Sampled based on Haldane equation | + | |Sampled based on Haldane equation. <br> '''Alternative:'''<math>0.063 \pm 0.028 </math> |
|<math> mM \times min^{-1} </math> | |<math> mM \times min^{-1} </math> | ||
| | | | ||
Revision as of 10:11, 29 May 2014
This enzyme splits fructose 1, 6-bisphosphate into two sugars that are isomers of each other. These two sugars are Dihydroxyacetone phosphate (DHAP) and Glyceraldehyde 3-phosphate (Gly3P).
Contents
Chemical equation
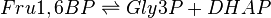
Rate equation
Reversible Uni-Bi Michaelis-Menten is used. [1]
![\frac{V_{mf} \frac{[Fru1,6BP]}{K_{Fru1,6BP}} - V_{mr}\frac{[DHAP][G3P]}{K_{DHAP}K_{Gly3P}} }{1 + \frac{[Fru1,6BP]}{K_{Fru1,6BP}} + \frac{[DHAP]}{K_{DHAP}} +\frac{[Gly3P]}{K_{Gly3P}} + \frac{[DHAP][Gly3P]}{K_{DHAP}K_{Gly3P}} }](/wiki/images/math/d/f/0/df0be717a9e6f06e44e264eacb312fd7.png)
Parameters
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
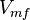
|
0.08 [2] | 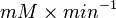
|
Hela cell line | |
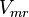
|
0.063[2] | 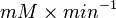
|
Rodent AS-30D hepatoma | |
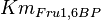
|
0.009[2] | mM | Hela cell line | |
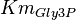
|
0.16[2] | mM | Rodent AS-30D hepatoma | |
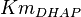
|
0.08[2] | mM | Rodent AS-30D hepatoma |
Parameters with uncertainty
- The value for
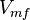 is collected from Hernandez et. al. [2]. The
is collected from Hernandez et. al. [2]. The 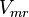 is calcualted from the sampled
is calcualted from the sampled 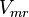 ,
, 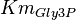 ,
, 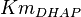 and
and  values using the Haldane equation. Alternatively the reported value from AS-30D can be considered. The standard deviation can be calculated based on the same ratio of
values using the Haldane equation. Alternatively the reported value from AS-30D can be considered. The standard deviation can be calculated based on the same ratio of 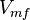 . However, as it is from a different species an extra 20% of uncertainty will be added. This gives the value
. However, as it is from a different species an extra 20% of uncertainty will be added. This gives the value 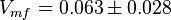
- The value for
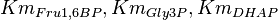 are collected from Ali D. Malay et. al. [3] for wildtype Aldolase B gene at
are collected from Ali D. Malay et. al. [3] for wildtype Aldolase B gene at 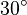 C.
C.
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
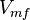
|
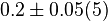 [2] [2]
|
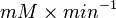
|
Hela cell line | |
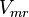
|
Sampled based on Haldane equation. Alternative: 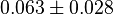
|
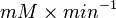
|
||
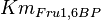
|
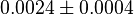
|
mM | Human cell | |
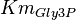
|
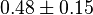
|
mM | Human cell | |
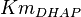
|
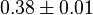
|
mM | Human cell |
Equilibrium constant
| Equilibrium constant | Conditions | Source |
|---|---|---|
| 0.10 | pH=7, T=25°C | Voet et al.[4] from Newshole et al. (1973) [5]p 97:
|
| 0.067 | pH=7, T=25°C | Lehninger, (1975)[6] p 407:
|
References
- ↑ Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 (doi)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Marín-Hernández A , Rodríguez-Enríquez S, Vital-González P A, et al. (2006). Determining and understanding the control of glycolysis in fast-growth tumor cells. Flux control by an over-expressed but strongly product-inhibited hexokinase. FEBS J., 273 , pp. 1975–1988(doi)
- ↑ Malay AD, Procious SL, Tolan DR. (2002). The temperature dependence of activity and structure for most prevalent mutant aldolase B associated with hereditary fructose intolerance, Arch BiochemBiophys 408: 295–304.
- ↑ Voet, D., Voet., J.G. and Pratt, C. W. (1999) Fundamentals of biochemistry, Wiley
- ↑ Newshole, E.A. and Stuart, C. (1973) Regulation in Metabolism, Wiley
- ↑ Lehninger, A.L. (1975) Biochemistry (2nd edn), Worth
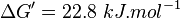 ,
, 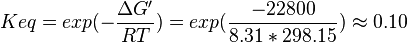
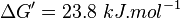 ,
,