Double-bond reductase (DBR)
Revision as of 15:45, 25 May 2016 by Aliah.hawari (talk | contribs) (Created page with "You can go back to main page of the kinetic model [http://www.systemsbiology.ls.manchester.ac.uk/wiki/index.php/Kinetic_Model_of_Monoterpenoid_Biosynthesis_Wiki here]. '''Leg...")
You can go back to main page of the kinetic model here.
Legend:
| Have not started ·· | 1 -2 data found ·· | 3-4 data found ·· | sufficient data found/estimated ·· | data distribution generated ·· | data sampled |
| to do | DONE! |
Contents
What we know
Issues
Strategies
Reaction catalysed
Enzyme and Metabolite Background Information
Long metabolite names are abbreviated in the model for clarity and standard identification purposes.
| Metabolite | Abbreviation | Chemical Formula | Molar mass (g/mol) | ChEBI | ChEMBL | PubChem | BRENDA | PlantCyc |
|---|---|---|---|---|---|---|---|---|
| limonene-3-hydroxylase | L3H | 56.1 kD | 1.14.13.47 | MONOMER-6761 | ||||
| (-)-4S-limonene | limonene | C10H16 | 136.24 | 15384 | 449062 | 22311 or 439250 | ||
| (-)-trans-isopiperitenol | isopiperitenol | C10H16O | 152.23344 | 15406 | 439410 | |||
| NADPH | C21H30N7O17P3 | 745.42116 | 16474 | |||||
| NADP+ | C21H29N7O17P3 | 744.41322 | 18009 | |||||
| Dioxygen | O2 | O2 | 31.99880 | 15379 | ||||
| water | H2O | H2O | 18.01530 | 15377 |
Equation Rate
Limonene-hydroxylase (L3H) is modelled using the reversible Michaelis-Menten equation.
| Parameter | Description | Reference |
|---|---|---|
| VL3H | Reaction rate for Limonene-3-hydroxylase | ref |
| Vmaxforward | Maximum reaction rate towards the production of (-)-trans-isopiperitenol | ref |
| Kmlimonene | Michaelis-Menten constant for Limonene | ref |
| Kmisopiperitenol | Michaelis-Menten constant for (-)-trans-isopiperitenol | ref |
| KmNADPH | Michaelis-Menten constant for NADPH | ref |
| KmNADP | Michaelis-Menten constant for NADP+ | ref |
| Keq | Equilibrium constant | ref |
| [limonene] | Limonene concentration | ref |
| [isopiperitenol] | (-)-trans-isopiperitenol concentration | ref |
| [NADPH] | NADPH concentration | ref |
| [NADP] | NADP+ concentration | ref |
Strategies for estimating the kinetic parameter values
Standard Gibbs Free energy
The Gibbs free energy for PGR is -3.9565125 kcal.mol^-1. This value is estimated from the 'Contribution group' method by Latendresse, M. and is available from MetaCyc (EC 1.3.1.81) [1].
Calculating the Equilibrium Constant
The equilibrium constant can be calculated using the Van't Hoff Isotherm equation:
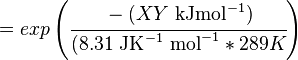
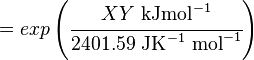
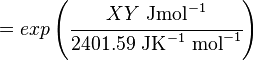
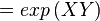
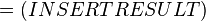
where;
| Keq | Equilibrium constant |
| -ΔG° | Gibbs free energy change. For (INSERT ENZYME) it is (INSERT VALUE) kJmol-1 |
| R | Gas constant with a value of 8.31 JK-1mol-1 |
| T | Temperature which is always expressed in kelvin |
Extracting Information from (INSERT SUBSTRATE/PRODUCT) Production Rates
A table will go here
Published Kinetic Parameter Values
A table will go here.
Detailed description of kinetic values obtained from literature
A more detailed description of the values listed above can be found here .
Simulations
References
- ↑ Latendresse M. (2013). "Computing Gibbs Free Energy of Compounds and Reactions in MetaCyc."
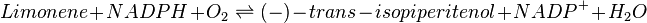
![V_\mathrm{L3H} = Vmax_\mathrm{forward} * \cfrac {\left ( \cfrac{[limonene]}{Km_\mathrm{limonene}} * \cfrac {[NADPH]}{Km_\mathrm{NADPH}} \right ) * \left ( 1 - \cfrac {[isopiperitenol]*[NADP]}{[limonene]*[NADPH]*K_\mathrm{eq}} \right )}
{ \left (1 + \cfrac {[NADPH]}{Km_\mathrm{NADPH}} + \cfrac {[NADP]}{Km_\mathrm{NADP}} \right ) + \left ( 1+ \cfrac {[limonene]}{Km_\mathrm{limonene}} + \cfrac {[isopiperitenol]}{Km_\mathrm{isopiperitenol}} \right ) }](/wiki/images/math/6/3/7/6372d5716522f3a6100b47a63f36e873.png)