Dehydrogenase
A dehydrogenase is an enzyme that oxidizes a substrate by a reduction reaction that transfers one or more hydrides (H−) to an electron acceptor, usually Nicotinamide adenine dinucleotide NAD+/NADP.
Contents
Chemical reaction
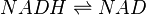
Rate equation
Reversible mass action rate law is used
![K_{1}[NADH] - K_{2}[NAD]](/wiki/images/math/5/7/f/57ff1f0db057e5957f43bd3cf4f50fd1.png)
Parameters
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
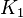
|
250 [1] | HeLa cell line | |
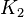
|
1[1] |
Parameters with uncertainty
- This is a pseudo-reaction, modelled using mass action kinetics (i.e., as non-saturable, non-enzymatic reactions) to maintain maximal compatibility with Pyridine Nucleotides balances. The parameter values were adjusted through model simulations and the values chosen were those that best predicted NAD+ concentration (determined experimentally) in Hernandez et. al. [1]. No information is available about the uncertainty of these parameters. As these parameters are strictly positive, they are sampled using a log-normal distribution as are and values. The means are set to the value using fitted by Hernandez et al. [1] for the fixed-parameter model. The sampling of the parameters are made to fall within
![[0.001\times mean \quad 1000 \times mean ]](/wiki/images/math/7/0/5/70565b13097c64cb12ee455eb2006ee7.png) to allow a large exploration of the parameter space. We use the Range rule to calculate the mean and standard deviation from maximum and minimum value. For both
to allow a large exploration of the parameter space. We use the Range rule to calculate the mean and standard deviation from maximum and minimum value. For both 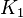 the minimum and maximum value lies between 0.25 and 250000. So the mean for
the minimum and maximum value lies between 0.25 and 250000. So the mean for 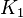 is 125000.125 and standard deviation would be 62500.0625 and for
is 125000.125 and standard deviation would be 62500.0625 and for 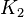 the minimum and maximum value lies between 0.001 and 1000. So the mean for
the minimum and maximum value lies between 0.001 and 1000. So the mean for 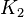 is 500.0005 and standard deviation would be 250.00025.
is 500.0005 and standard deviation would be 250.00025.
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
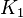
|
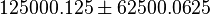
|
HeLa cell line | |
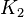
|
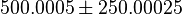
|