Difference between revisions of "Degradation of r"
(→Parameters) |
(→Parameters) |
||
| Line 25: | Line 25: | ||
|- | |- | ||
|<math>d_{mR}</math> | |<math>d_{mR}</math> | ||
| − | |<math>0. | + | |<math>0.03-0.4</math> <ref name="Kristoffersen2012"> [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3446304/pdf/gb-2012-13-4-r30.pdf Kristoffersen SM, Haase C, Weil MR, et al. ''Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium.'' Genome Biology. 2012;13(4):R30.]</ref> <ref name="Ortiz1984"> [http://www.sciencedirect.com/science/book/9780125286206 Ortiz-Ortiz, L. and Bojalil, L.F. and Yakoleff, V. ''Biological, Biochemical, and Biomedical Aspects of Actinomycetes.'' Elsevier, 1984]</ref> |
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
|<math>0.0018 s^{-1}</math><ref name="Mehra2008"></ref> | |<math>0.0018 s^{-1}</math><ref name="Mehra2008"></ref> | ||
| Line 40: | Line 40: | ||
Additionally, in a book on biological and biochemical aspects of ''Actinomycetes'', Ortiz-Ortiz et al. reported a range of mRNA degradation rates for ''Streptomyces antibioticus'' spores between <math>4-22.5 min </math>. By the same calculations, the corresponding mRNA degradation rate constants are in the range <math> 0.031-0.17 min^{-1} </math>. | Additionally, in a book on biological and biochemical aspects of ''Actinomycetes'', Ortiz-Ortiz et al. reported a range of mRNA degradation rates for ''Streptomyces antibioticus'' spores between <math>4-22.5 min </math>. By the same calculations, the corresponding mRNA degradation rate constants are in the range <math> 0.031-0.17 min^{-1} </math>. | ||
[[Image:DmR-text2.png|center|thumb|500px|Ortiz-Ortiz et al. 1984<ref name="Ortiz1984"></ref>]] | [[Image:DmR-text2.png|center|thumb|500px|Ortiz-Ortiz et al. 1984<ref name="Ortiz1984"></ref>]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
Revision as of 06:40, 16 October 2015
The mRNA of scbR (r) degrades.
Contents
Chemical equation
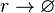
Rate equation
![r= d_{mR}\cdot[r]](/wiki/images/math/e/f/b/efba4e6bdd0c5b9124a5b33c3bfb3bba.png)
Parameters
The parameter of this reaction is the degradation rate of r (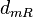 ). The parameter values were derived from calculations of the mRNA half life in gram positive bacteria with low GC content and in Actinomycetes.
). The parameter values were derived from calculations of the mRNA half life in gram positive bacteria with low GC content and in Actinomycetes.
| Name | Value | Units | Value in previous GBL models [1] [2] | Remarks-Reference |
|---|---|---|---|---|
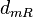
|
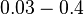 [3] [4] [3] [4]
|

|
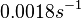 [1] [1]
(Range tested: (Bistability range: and |
In a study on global analysis of mRNA decay in gram positive bacteria by RNA-Seq, Kristoffersen et al. reported a range of values for the half-life (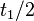 ) of mRNA in B. cereus between ) of mRNA in B. cereus between 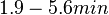 , with a median of , with a median of 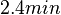 . From these values we calculated the mRNA degradation rate of scbR as per . From these values we calculated the mRNA degradation rate of scbR as per 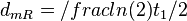 , which resulted in a degradation rate constant value range , which resulted in a degradation rate constant value range 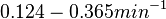 and a median and a median 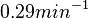 . .
 Kristoffersen et al. 2012[3] Additionally, in a book on biological and biochemical aspects of Actinomycetes, Ortiz-Ortiz et al. reported a range of mRNA degradation rates for Streptomyces antibioticus spores between 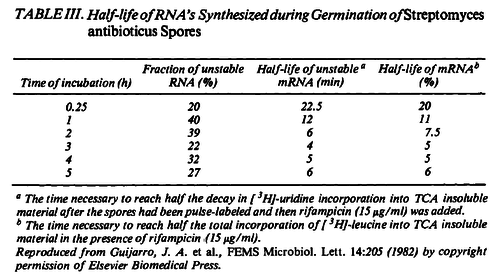 Ortiz-Ortiz et al. 1984[4] |
Parameters with uncertainty
The most plausible parameter value for the 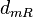 is decided to be
is decided to be 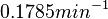 and the confidence interval
and the confidence interval 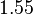 . This means that the mode of the PDF is 0.1785 and the range where 95% of the values are found is between
. This means that the mode of the PDF is 0.1785 and the range where 95% of the values are found is between 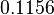 and
and 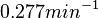 .
.
The probability distribution for the parameter, adjusted accordingly in order to reflect the above values, is the following:
The location and scale parameters of the distribution are:
| Parameter | μ | σ |
|---|---|---|
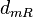
|
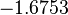
|
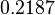
|
References
- ↑ 1.0 1.1 1.2 S. Mehra, S. Charaniya, E. Takano, and W.-S. Hu. A bistable gene switch for antibiotic biosynthesis: The butyrolactone regulon in streptomyces coelicolor. PLoS ONE, 3(7), 2008.
- ↑ 2.0 2.1 2.2 A. Chatterjee, L. Drews, S. Mehra, E. Takano, Y.N. Kaznessis, and W.-S. Hu. Convergent transcription in the butyrolactone regulon in streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS ONE, 6(7), 2011.
- ↑ 3.0 3.1 Kristoffersen SM, Haase C, Weil MR, et al. Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium. Genome Biology. 2012;13(4):R30.
- ↑ 4.0 4.1 Ortiz-Ortiz, L. and Bojalil, L.F. and Yakoleff, V. Biological, Biochemical, and Biomedical Aspects of Actinomycetes. Elsevier, 1984


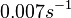
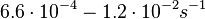 )
)
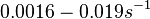
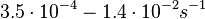
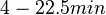 . By the same calculations, the corresponding mRNA degradation rate constants are in the range
. By the same calculations, the corresponding mRNA degradation rate constants are in the range 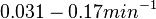 .
.