Difference between revisions of "Degradation of r"
(→Parameters with uncertainty) |
(→Parameters with uncertainty) |
||
| Line 34: | Line 34: | ||
==Parameters with uncertainty== | ==Parameters with uncertainty== | ||
| − | The most plausible parameter value for the <math>d_{mR}</math> is decided to be <math> 0.1785 min^{-1} </math> and the confidence interval <math> 1.55 </math>. This means that the mode of the PDF is 0.1785 and the range where 95% of the values are found is between 0. | + | The most plausible parameter value for the <math>d_{mR}</math> is decided to be <math> 0.1785 min^{-1} </math> and the confidence interval <math> 1.55 </math>. This means that the mode of the PDF is 0.1785 and the range where 95% of the values are found is between <math>0.1156</math> and <math>0.277 min^{-1}</math>. |
The probability distributionfor the parameter, adjusted accordingly in order to reflect the above values, is the following: | The probability distributionfor the parameter, adjusted accordingly in order to reflect the above values, is the following: | ||
Revision as of 21:00, 28 September 2015
The mRNA of ScbR (r) degrades.
Contents
Chemical equation
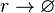
Rate equation
![r= d_{mR}\cdot[r]](/wiki/images/math/e/f/b/efba4e6bdd0c5b9124a5b33c3bfb3bba.png)
Parameters
The parameter of this reaction is the degradation rate of r (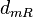 ).
).
| Name | Value | Units | Origin | Remarks |
|---|---|---|---|---|
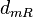
|
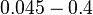 [1] [2] [1] [2]
|

|
Half life of mRNA in Gram positive bacteria (low GC content)
Half life of mRNA in Actinomycetes |
Parameters with uncertainty
The most plausible parameter value for the 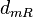 is decided to be
is decided to be 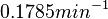 and the confidence interval
and the confidence interval 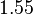 . This means that the mode of the PDF is 0.1785 and the range where 95% of the values are found is between
. This means that the mode of the PDF is 0.1785 and the range where 95% of the values are found is between 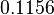 and
and 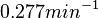 .
.
The probability distributionfor the parameter, adjusted accordingly in order to reflect the above values, is the following:
The location and scale parameters of the distribution are:
| Parameter | μ | σ |
|---|---|---|
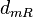
|
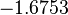
|
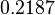
|
References
- ↑ Kristoffersen SM, Haase C, Weil MR, et al. Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium. Genome Biology. 2012;13(4):R30.
- ↑ Ortiz-Ortiz, L. and Bojalil, L.F. and Yakoleff, V. Biological, Biochemical, and Biomedical Aspects of Actinomycetes. Elsevier, 2013