Difference between revisions of "Degradation of R2"
(→Parameters with uncertainty) |
|||
| Line 34: | Line 34: | ||
==Parameters with uncertainty== | ==Parameters with uncertainty== | ||
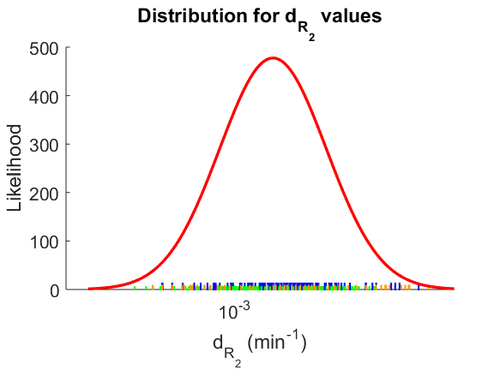
| − | The most plausible parameter value for the <math>d_{ | + | The most plausible parameter value for the <math>d_{R_{2}}</math> is decided to be <math> 0.00118 min^{-1} </math> and the confidence interval <math> 1.41 </math>. This means that the mode of the PDF is 0.00118 and the range where 95% of the values are found is between <math>0.8369\cdot 10^{-3}</math> and <math>1.67 \cdot 10^{-3}min^{-1}</math>. |
The probability distribution for the parameter, adjusted accordingly in order to reflect the above values, is the following: | The probability distribution for the parameter, adjusted accordingly in order to reflect the above values, is the following: | ||
| − | [[Image: | + | [[Image:DR2.png|500px]] |
The location and scale parameters of the distribution are: | The location and scale parameters of the distribution are: | ||
Revision as of 03:26, 2 October 2015
The ScbR homo-dimer (R2) degrades.
Contents
Chemical equation
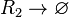
Rate equation
![r= d_{R_{2}}\cdot[R_{2}]](/wiki/images/math/c/5/e/c5e7cd8f67a62c67acc417614bcee8ce.png)
Parameters
The parameter of this reaction is the degradation rate of R2 (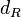 ).
).
| Name | Value | Units | Origin | Remarks |
|---|---|---|---|---|
|
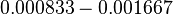 [1] [1]
|

|
Degradation rates of proteins in S. coelicolor
based on radioactive decay |
Parameters with uncertainty
The most plausible parameter value for the 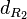 is decided to be
is decided to be 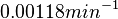 and the confidence interval
and the confidence interval 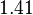 . This means that the mode of the PDF is 0.00118 and the range where 95% of the values are found is between
. This means that the mode of the PDF is 0.00118 and the range where 95% of the values are found is between 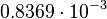 and
and 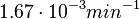 .
.
The probability distribution for the parameter, adjusted accordingly in order to reflect the above values, is the following:
The location and scale parameters of the distribution are:
| Parameter | μ | σ |
|---|---|---|
|
|
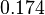
|
References
- ↑ Trötschel C, Albaum SP, Poetsch A. Proteome turnover in bacteria: current status for Corynebacterium glutamicum and related bacteria. Microbial biotechnology. 2013;6(6):708-719.