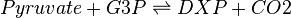Difference between revisions of "DXS"
Aliah.hawari (talk | contribs) (→DXS parameters) |
Aliah.hawari (talk | contribs) (→DXS parameters) |
||
| Line 35: | Line 35: | ||
| Kcat || Forward || DXS || 229.7 || 1/min || 16 || from E. coli wild type DXS, with non-optimal buffer: 40mM Tris, pH 8, 37C ; Km_GAP:52.5 +/- 8.3 microM; Km_pyruvate: 86.3 +/- 16.2 microM, kcat: 145.50 +/- 12.7 1/min || <ref name="Brammer2011"> [https://www.ncbi.nlm.nih.gov/pubmed/21878632 Brammer, L.A. 2011] "1-FDeoxy-D-xylulose 5-phosphate synthase catalyzes a novel random sequential mechanism", JBioChem, '''283'''(42):36522-36531.</ref> | | Kcat || Forward || DXS || 229.7 || 1/min || 16 || from E. coli wild type DXS, with non-optimal buffer: 40mM Tris, pH 8, 37C ; Km_GAP:52.5 +/- 8.3 microM; Km_pyruvate: 86.3 +/- 16.2 microM, kcat: 145.50 +/- 12.7 1/min || <ref name="Brammer2011"> [https://www.ncbi.nlm.nih.gov/pubmed/21878632 Brammer, L.A. 2011] "1-FDeoxy-D-xylulose 5-phosphate synthase catalyzes a novel random sequential mechanism", JBioChem, '''283'''(42):36522-36531.</ref> | ||
|- | |- | ||
| − | | Kcat || Forward || DXS || 153.6 || 1/min || 16 || from E. coli wild type DXS, with non-optimal buffer: 40mM Tris, pH 8, 37C ; Km_GAP:52.5 +/- 8.3 microM; Km_pyruvate: 86.3 +/- 16.2 microM, kcat: 145.50 +/- 12.7 1/min || <ref> | + | | Kcat || Forward || DXS || 153.6 || 1/min || 16 || from E. coli wild type DXS, with non-optimal buffer: 40mM Tris, pH 8, 37C ; Km_GAP:52.5 +/- 8.3 microM; Km_pyruvate: 86.3 +/- 16.2 microM, kcat: 145.50 +/- 12.7 1/min || <ref name="Brammer2011"></ref> |
|- | |- | ||
| − | | Kcat || Forward || DXS || 145.5 || 1/min || 16 || from E. coli wild type DXS, with non-optimal buffer: 40mM Tris, pH 8, 37C ; Km_GAP:52.5 +/- 8.3 microM; Km_pyruvate: 86.3 +/- 16.2 microM, kcat: 145.50 +/- 12.7 1/min || <ref> | + | | Kcat || Forward || DXS || 145.5 || 1/min || 16 || from E. coli wild type DXS, with non-optimal buffer: 40mM Tris, pH 8, 37C ; Km_GAP:52.5 +/- 8.3 microM; Km_pyruvate: 86.3 +/- 16.2 microM, kcat: 145.50 +/- 12.7 1/min || <ref name="Brammer2011"></ref> |
|- | |- | ||
| − | | Kcat || Forward || DXS || 209 || 1/min || 16 || from E. coli wild type DXS, with non-optimal buffer: 100mM Tris, pH 8, 37C ; Km_GAP:279.0+/- 7.2microM; Km_pyruvate: 74.70+/- 7.3 microM, kcat: 209.0 +/- 6.3 1/min || <ref> | + | | Kcat || Forward || DXS || 209 || 1/min || 16 || from E. coli wild type DXS, with non-optimal buffer: 100mM Tris, pH 8, 37C ; Km_GAP:279.0+/- 7.2microM; Km_pyruvate: 74.70+/- 7.3 microM, kcat: 209.0 +/- 6.3 1/min || <ref name="Brammer2011"></ref> |
|- | |- | ||
| − | | Kcat || Forward || DXS || 173 || 1/min || 16 || from E. coli wild type DXS, with non-optimal buffer: 100mM Tris, pH 8, 37C ; Km_GAP:279.0+/- 7.2microM; Km_pyruvate: 74.70+/- 7.3 microM, kcat: 209.0 +/- 6.3 1/min || <ref> | + | | Kcat || Forward || DXS || 173 || 1/min || 16 || from E. coli wild type DXS, with non-optimal buffer: 100mM Tris, pH 8, 37C ; Km_GAP:279.0+/- 7.2microM; Km_pyruvate: 74.70+/- 7.3 microM, kcat: 209.0 +/- 6.3 1/min || <ref name="Brammer2011"></ref> |
|- | |- | ||
| − | | Kcat || Forward || DXS || 246 || 1/min || 16 || from E. coli wild type DXS, with non-optimal buffer: 100mM Tris, pH 8, 37C ; Km_GAP:279.0+/- 7.2microM; Km_pyruvate: 74.70+/- 7.3 microM, kcat: 209.0 +/- 6.3 1/min || <ref> | + | | Kcat || Forward || DXS || 246 || 1/min || 16 || from E. coli wild type DXS, with non-optimal buffer: 100mM Tris, pH 8, 37C ; Km_GAP:279.0+/- 7.2microM; Km_pyruvate: 74.70+/- 7.3 microM, kcat: 209.0 +/- 6.3 1/min || <ref name="Brammer2011"></ref> |
|- | |- | ||
| − | | Kcat || Forward || GAP || 48 || 1/min || 256 || Taken from Cane 2001's ref20. E.coli DXS in 40mM Tris, pH7.5, 37¡C. Km pyruvate 2.9 ± 0.5 mM. || Boronat1999 | + | | Kcat || Forward || GAP || 48 || 1/min || 256 || Taken from Cane 2001's ref20. E.coli DXS in 40mM Tris, pH7.5, 37¡C. Km pyruvate 2.9 ± 0.5 mM. ||<ref name="Boronat1999"> [https://www.ncbi.nlm.nih.gov/pubmed/21878632 Brammer, L.A. 2011] "1-Deoxy-D-xylulose 5-phosphate synthase catalyzes a novel random sequential mechanism", JBioChem, '''283'''(42):36522-36531.</ref> |
|- | |- | ||
| − | | Kcat || Forward || GAP || 66 || 1/min || 128 || DXPS2; in vitro- S. coelicolor gene expressed in E. coli; pH 7.5, 47C. || Cane2001 | + | | Kcat || Forward || GAP || 66 || 1/min || 128 || DXPS2; in vitro- S. coelicolor gene expressed in E. coli; pH 7.5, 47C. || <ref name="Cane2001"> [https://www.ncbi.nlm.nih.gov/pubmed/11408165 Cane, D.E., ''et. al.''. 2001] "Molecular cloning, expression and characterization of the first three genes in the mevalonate-independent isoprenoid pathway in ''Streptomyces coelicolor'' ", Bioorganic & Medicinal Chemistry, '''9''':1467-1477.</ref> |
|- | |- | ||
| − | | Kcat || Forward || GAP || 114 || 1/min || 8 || from R. capsulatus, pH 7.4, 37C || Eubanks2003, | + | | Kcat || Forward || GAP || 114 || 1/min || 8 || from R. capsulatus, pH 7.4, 37C || <ref name="Eubanks2003"> [http://pubs.acs.org/doi/abs/10.1021/bi0205303 Eubanks, L.M. & Poulter, C.D. 2003.] "Rhodobacter capsulatus 1-Deoxy-D-xylulose 5-Phosphate Synthase: Steady-State Kinetics and Substrate Binding† ", Biochemistry, '''42''':1140-1149.</ref> |
|- | |- | ||
| − | | Kcat || Forward || GAP || 660 || 1/min || 64 || from Plasmodium, expressed in E. coli. Look at Table 3. pH7-7.5;37C, Km_GAp:19 +/- 4 microM; Km_Pyruvate: 870 +/- 110 microM. || Handa2013, | + | | Kcat || Forward || GAP || 660 || 1/min || 64 || from Plasmodium, expressed in E. coli. Look at Table 3. pH7-7.5;37C, Km_GAp:19 +/- 4 microM; Km_Pyruvate: 870 +/- 110 microM. || <ref name="Handa2013"> [http://onlinelibrary.wiley.com/doi/10.1016/j.fob.2013.01.007/full Handa, S. ''et. al.'' 2013.] "Production of recombinant 1-deoxy-d-xylulose-5-phosphate synthase from Plasmodium vivax in Escherichia coli", Biochemistry, '''3''':124-129.</ref> |
|- | |- | ||
| − | | Kcat || Forward || GAP || 1608 || 1/min || 8 || dxs11 from Agrobacterium tumefaciens, pH8.0, 37¡C, expressed in E. coli || Lee2007, | + | | Kcat || Forward || GAP || 1608 || 1/min || 8 || dxs11 from Agrobacterium tumefaciens, pH8.0, 37¡C, expressed in E. coli || <ref name="Lee2007"> [http://www.sciencedirect.com/science/article/pii/S0168165606009667 Lee, J. '' et. al.'' 2007.] "Cloning and characterization of the dxs gene, encoding 1-deoxy-d-xylulose 5-phosphate synthase from Agrobacterium tumefaciens, and its overexpression in Agrobacterium tumefaciens", JBiotech, '''128''':555-566.</ref> |
|- | |- | ||
| − | | Kcat || Forward || GAP || 120 || 1/min || 64 || from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM || Matsushima2012, | + | | Kcat || Forward || GAP || 120 || 1/min || 64 || from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM || <ref name="Matsushima2012"> [https://www.ncbi.nlm.nih.gov/pubmed/22325894 Matsushima, D. ''et. al''2012.] "The single cellular green microalga Botryococcus braunii, race B possesses three |
| + | distinct 1-deoxy-d-xylulose 5-phosphate synthases", PlantSci, 185-186:309-320.</ref> | ||
|- | |- | ||
| − | | Kcat || Forward || GAP || 120 || 1/min || 64 || from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM || Matsushima2012 | + | | Kcat || Forward || GAP || 120 || 1/min || 64 || from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM || <ref name='Matsushima2012></ref> |
|- | |- | ||
| − | | Kcat || Forward || GAP || 360 || 1/min || 64 || from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM || Matsushima2012 | + | | Kcat || Forward || GAP || 360 || 1/min || 64 || from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM || <ref name='Matsushima2012></ref> |
|- | |- | ||
| − | | Kcat || Forward || Pyruvate || 570 || 1/min || 64 || from Plasmodium, expressed in E. coli. Look at Table 3. pH7-7.5;37C, Km_GAp:19 +/- 4 microM; Km_Pyruvate: 870 +/- 110 microM. || Handa2013 | + | | Kcat || Forward || Pyruvate || 570 || 1/min || 64 || from Plasmodium, expressed in E. coli. Look at Table 3. pH7-7.5;37C, Km_GAp:19 +/- 4 microM; Km_Pyruvate: 870 +/- 110 microM. || <ref name="Handa2013"> </ref> |
|- | |- | ||
| − | | Kcat || Forward || Pyruvate || 144 || 1/min || 64 || from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM || Matsushima2012 | + | | Kcat || Forward || Pyruvate || 144 || 1/min || 64 || from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM || <ref name='Matsushima2012></ref> |
|- | |- | ||
| − | | Kcat || Forward || Pyruvate || 114 || 1/min || 64 || from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM || Matsushima2012 | + | | Kcat || Forward || Pyruvate || 114 || 1/min || 64 || from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM || <ref name='Matsushima2012></ref> |
|- | |- | ||
| − | | Kcat || Forward || Pyruvate || 312 || 1/min || 64 || from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM || Matsushima2012 | + | | Kcat || Forward || Pyruvate || 312 || 1/min || 64 || from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM || <ref name='Matsushima2012></ref> |
|} | |} | ||
Revision as of 14:42, 23 March 2017
You can go back to main page of the kinetic model here.
The DXS reaction (EC 2.2.1.7)
Deoxyxylulose-5-phosphate synthase (DXS) catalyses the production of 1-deoxy-D-xylulose 5-phosphate (DXP) from pyruvate and glyceraldehyde 3-phosphate (G3P). This reaction is the first step in the MEP pathway.
Modelling DXS
In the kinetic model, the DXS reaction is modelled with reversible Michaelis-Menten using the Hanekom [1] bi-bi random order generic equation. In total, this reaction requires five kinetic parameters (Kms for all substrates and products, and a forward Kcat) and one thermodynamic parameter (Equilibrium constant, Keq).
![V_\mathrm{DXS}= \cfrac{Kcat_\mathrm{forward} \bullet [DXS] \bullet \left( \cfrac{[Pyr]}{Km_\mathrm{DXS}} \right) \bullet \left( \cfrac{[G3P]}{Km_\mathrm{g3p}} \right) \bullet \left( 1 - \cfrac{\left( \cfrac{[DXP]\bullet[CO2]}{[Pyr]\bullet[G3P]} \right)}{K_\mathrm{eq}} \right)} {\left( 1 + \cfrac {[Pyr]}{Km_\mathrm{pyr}} + \cfrac{[CO2]}{Km_\mathrm{co2}}\right) \bullet \left( 1 + \cfrac{[G3P]}{Km_\mathrm{g3p}} + \cfrac{[DXP]}{KM_\mathrm{dxp}} \right)}](/wiki/images/math/4/a/8/4a8faa589ce739bde2b441190759da1b.png)
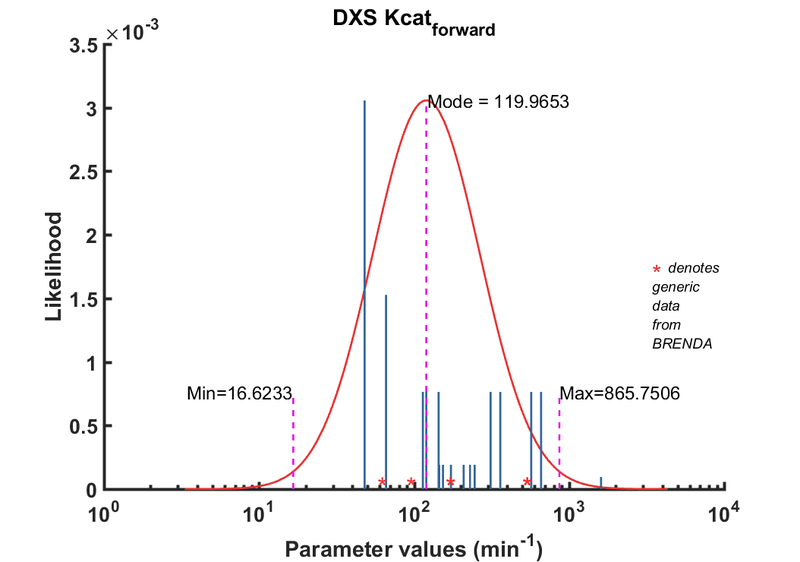
DXS parameters
| Parameter | Direction | Substrate | Value | Unit | Weight | Description | Reference |
|---|---|---|---|---|---|---|---|
| Kcat | Forward | DXS | 229.7 | 1/min | 16 | from E. coli wild type DXS, with non-optimal buffer: 40mM Tris, pH 8, 37C ; Km_GAP:52.5 +/- 8.3 microM; Km_pyruvate: 86.3 +/- 16.2 microM, kcat: 145.50 +/- 12.7 1/min | [2] |
| Kcat | Forward | DXS | 153.6 | 1/min | 16 | from E. coli wild type DXS, with non-optimal buffer: 40mM Tris, pH 8, 37C ; Km_GAP:52.5 +/- 8.3 microM; Km_pyruvate: 86.3 +/- 16.2 microM, kcat: 145.50 +/- 12.7 1/min | [2] |
| Kcat | Forward | DXS | 145.5 | 1/min | 16 | from E. coli wild type DXS, with non-optimal buffer: 40mM Tris, pH 8, 37C ; Km_GAP:52.5 +/- 8.3 microM; Km_pyruvate: 86.3 +/- 16.2 microM, kcat: 145.50 +/- 12.7 1/min | [2] |
| Kcat | Forward | DXS | 209 | 1/min | 16 | from E. coli wild type DXS, with non-optimal buffer: 100mM Tris, pH 8, 37C ; Km_GAP:279.0+/- 7.2microM; Km_pyruvate: 74.70+/- 7.3 microM, kcat: 209.0 +/- 6.3 1/min | [2] |
| Kcat | Forward | DXS | 173 | 1/min | 16 | from E. coli wild type DXS, with non-optimal buffer: 100mM Tris, pH 8, 37C ; Km_GAP:279.0+/- 7.2microM; Km_pyruvate: 74.70+/- 7.3 microM, kcat: 209.0 +/- 6.3 1/min | [2] |
| Kcat | Forward | DXS | 246 | 1/min | 16 | from E. coli wild type DXS, with non-optimal buffer: 100mM Tris, pH 8, 37C ; Km_GAP:279.0+/- 7.2microM; Km_pyruvate: 74.70+/- 7.3 microM, kcat: 209.0 +/- 6.3 1/min | [2] |
| Kcat | Forward | GAP | 48 | 1/min | 256 | Taken from Cane 2001's ref20. E.coli DXS in 40mM Tris, pH7.5, 37¡C. Km pyruvate 2.9 ± 0.5 mM. | [3] |
| Kcat | Forward | GAP | 66 | 1/min | 128 | DXPS2; in vitro- S. coelicolor gene expressed in E. coli; pH 7.5, 47C. | [4] |
| Kcat | Forward | GAP | 114 | 1/min | 8 | from R. capsulatus, pH 7.4, 37C | [5] |
| Kcat | Forward | GAP | 660 | 1/min | 64 | from Plasmodium, expressed in E. coli. Look at Table 3. pH7-7.5;37C, Km_GAp:19 +/- 4 microM; Km_Pyruvate: 870 +/- 110 microM. | [6] |
| Kcat | Forward | GAP | 1608 | 1/min | 8 | dxs11 from Agrobacterium tumefaciens, pH8.0, 37¡C, expressed in E. coli | [7] |
| Kcat | Forward | GAP | 120 | 1/min | 64 | from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM | [8] |
| Kcat | Forward | GAP | 120 | 1/min | 64 | from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM | [8] |
| Kcat | Forward | GAP | 360 | 1/min | 64 | from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM | [8] |
| Kcat | Forward | Pyruvate | 570 | 1/min | 64 | from Plasmodium, expressed in E. coli. Look at Table 3. pH7-7.5;37C, Km_GAp:19 +/- 4 microM; Km_Pyruvate: 870 +/- 110 microM. | [6] |
| Kcat | Forward | Pyruvate | 144 | 1/min | 64 | from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM | [8] |
| Kcat | Forward | Pyruvate | 114 | 1/min | 64 | from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM | [8] |
| Kcat | Forward | Pyruvate | 312 | 1/min | 64 | from Botrycoccus braunnii. Three recombinant enzymes used: DXS-I, DXS-II, DXS-III which are different by the digestion pattern using Xhol and BamHI. expressed in E. coli; pH 7.8 , 32 C; Km 1800 +/- 200 microM | [8] |
References
- ↑ Hanekom, A. J. 2006. "Generic kinetic equations for modelling multisubstrate reactions in computational systems biology", MSc Thesis submitted at the University of Stellenbosch
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Brammer, L.A. 2011 "1-FDeoxy-D-xylulose 5-phosphate synthase catalyzes a novel random sequential mechanism", JBioChem, 283(42):36522-36531.
- ↑ Brammer, L.A. 2011 "1-Deoxy-D-xylulose 5-phosphate synthase catalyzes a novel random sequential mechanism", JBioChem, 283(42):36522-36531.
- ↑ Cane, D.E., et. al.. 2001 "Molecular cloning, expression and characterization of the first three genes in the mevalonate-independent isoprenoid pathway in Streptomyces coelicolor ", Bioorganic & Medicinal Chemistry, 9:1467-1477.
- ↑ Eubanks, L.M. & Poulter, C.D. 2003. "Rhodobacter capsulatus 1-Deoxy-D-xylulose 5-Phosphate Synthase: Steady-State Kinetics and Substrate Binding† ", Biochemistry, 42:1140-1149.
- ↑ 6.0 6.1 Handa, S. et. al. 2013. "Production of recombinant 1-deoxy-d-xylulose-5-phosphate synthase from Plasmodium vivax in Escherichia coli", Biochemistry, 3:124-129. Cite error: Invalid
<ref>tag; name "Handa2013" defined multiple times with different content - ↑ Lee, J. et. al. 2007. "Cloning and characterization of the dxs gene, encoding 1-deoxy-d-xylulose 5-phosphate synthase from Agrobacterium tumefaciens, and its overexpression in Agrobacterium tumefaciens", JBiotech, 128:555-566.
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 Matsushima, D. et. al2012. "The single cellular green microalga Botryococcus braunii, race B possesses three distinct 1-deoxy-d-xylulose 5-phosphate synthases", PlantSci, 185-186:309-320.