DXR
You can go back to main page of the kinetic model here.
DXR reaction
The 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR, EC 1.1.1.267) is the second step in the MEP pathway that catalyses the production of 2-C-methyl-D-erythritol 4-phosphate (MEP) from 1-deoxy-D-xylulose 5-phosphate (DXP).
Modelling DXR
DXR is modelled reversible with the Michaelis-Menten rate law using Hanekom's generic random order bi-bi equation [1] [2]. There are a total of five kinetic parameters (2 forward Kms, 2 reverse Kms and 1 Kcat), and one thermodynamic parameter (Equilibrium constant , Keq).
DXR parameters
| Parameter | Direction | Substrate | Value | Unit | SEM | Weight | Description | Reference |
|---|---|---|---|---|---|---|---|---|
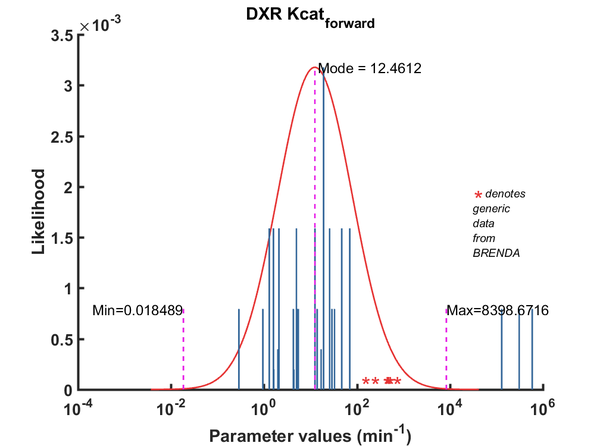
| Kcat | Forward | DXP | 70.3 | 1/min | NaN | 64 | from Mycobacterium tuberculosis, expressed in E. coli, pH 7.9, 30C | Dhiman2005, Br674260 |
| Kcat | Forward | DXP | 1740 | 1/min | 1 | 32 | from E. coli, pH 7.6, 37C, recombinant enzyme | Fox2005, BR672040 |
| Kcat | Forward | DXP | 318 | 1/min | NaN | 32 | Gene MtDXRfrom Mycobacterium tuberculosis, room temperature (20-22C), pH7.5 | Henriksson2006, Br671075 |
| Kcat | Forward | DXP | 120 | 1/min | 0.09 | 16 | gene from Francisella tularensis, expressed in E. coli, pH8, 22C | Jawaid2009, Br713353 |
| Kcat | Forward | DXP | 315 | 1/min | 0.19 | 32 | gene MtDXR from Mycobacterium tuberculosis, expressed in E. coli, pH 7.5, 25C. Experimental result, see Table 1. | Liu2012, BR724346 |
| Kcat | Forward | DXP | 1980 | 1/min | NaN | 32 | from E. coli, pH7.6, 37C | Walker2005 |
| Kcat | Reverse | DXR | 78 | 1/min | 0.1 | 64 | from Mycobacterium tuberculosis, transformed into competent E. coli BL21 (DE3) cells. pH7.5, 25C, using three different cofactors Mg2+, Mn2+ and Co2+. And NADPH /NADH BRENDA_lit: 654748, | Argyrou2004, Br654748 |
| Kcat | Forward | DXR | 126 | 1/min | 0.1 | 64 | from Mycobacterium tuberculosis, transformed into competent E. coli BL21 (DE3) cells. pH7.5, 25C, using three different cofactors Mg2+, Mn2+ and Co2+. And NADPH /NADH BRENDA_lit: 654748, | Argyrou2004, Br654748 |
| Kcat | Forward | DXR | 300 | 1/min | 0.1 | 64 | from Mycobacterium tuberculosis, transformed into competent E. coli BL21 (DE3) cells. pH7.5, 25C, using three different cofactors Mg2+, Mn2+ and Co2+. And NADPH /NADH BRENDA_lit: 654748, | Argyrou2004, Br654748 |
| Kcat | Forward | DXR | 96 | 1/min | 0.1 | 64 | from Mycobacterium tuberculosis, transformed into competent E. coli BL21 (DE3) cells. pH7.5, 25C, using three different cofactors Mg2+, Mn2+ and Co2+. And NADPH /NADH BRENDA_lit: 654748, | Argyrou2004, Br654748 |
| Kcat | Forward | DXR | 1152 | 1/min | 0.02 | 128 | DXPS1; in vitro- S. coelicolor gene expressed in E. coli; pH 7.5, 47C. | Cane2001 |
| Kcat | Forward | DXR | 1020 | 1/min | NaN | 16 | dxr gene from Synechocystis sp. PCC6803 cloned into E. coli. pH 7.8, 37C, BRENDA_lit:671590 | Fernandes2005, Br671590 |
| Kcat | Forward | DXR | 264 | 1/min | NaN | 8 | The ispC gene from Arabidopsis thaliana, expressed in E. coli, pH 8.0, 37C | Rohdich2006, Br673562 |
| Kcat | Reverse | DXR | 1.63 | 1/min | NaN | 8 | The ispC gene from Arabidopsis thaliana, expressed in E. coli, pH 8.0, 37C | Rohdich2006, Br673562 |
| Kcat | Forward | DXR | 17.4 | 1/min | NaN | 32 | 50C. from Thermotoga maritima. Kinetics were measured at different temperature: 50C and 85C, expressed in E. coli, pH 7.5, 50C. | Takenoya2010, Br712892 |
| Kcat | Forward | DXR | 330 | 1/min | NaN | 32 | 85C. from Thermotoga maritima. Kinetics were measured at different temperature: 50C and 85C, expressed in E. coli, pH 7.5, 50C. | Takenoya2010, Br712892 |
| Kcat | Forward | DXR | 840 | 1/min | NaN | 32 | from Synechocystis sp. PCC6803, expressed in E. coli, pH7.5, 37C, with three metal ions, Mn2+, Mg2+, Co2+ | Yin2003, Br654877 |
| Kcat | Forward | DXR | 300 | 1/min | NaN | 32 | from Synechocystis sp. PCC6803, expressed in E. coli, pH7.5, 37C, with three metal ions, Mn2+, Mg2+, Co2+ | Yin2003, Br654877 |
| Kcat | Forward | DXR | 258 | 1/min | NaN | 32 | from Synechocystis sp. PCC6803, expressed in E. coli, pH7.5, 37C, with three metal ions, Mn2+, Mg2+, Co2+ | Yin2003, Br654877 |
| Kcat | Reverse | MEP | 12.5 | 1/min | NaN | 64 | from Mycobacterium tuberculosis, expressed in E. coli, pH 7.9, 30C | Dhiman2005, Br674260 |
| Kcat | Reverse | MEP | 57 | 1/min | 0.06 | 32 | gene MtDXR from Mycobacterium tuberculosis, expressed in E. coli, pH 7.5, 25C. Experimental result, see Table 1. | Liu2012, BR724346 |
| Kcat | Reverse | NADP | 25.9 | 1/min | NaN | 64 | from Mycobacterium tuberculosis, expressed in E. coli, pH 7.9, 30C | Dhiman2005, Br674260 |
| Kcat | Forward | NADPH | 47.3 | 1/min | NaN | 64 | from Mycobacterium tuberculosis, expressed in E. coli, pH 7.9, 30C | Dhiman2005, Br674260 |
| Kcat | Forward | NADPH | 78 | 1/min | 0.04 | 16 | gene from Francisella tularensis, expressed in E. coli, pH8, 22C | Jawaid2009, Br713353 |
| Kcat | Forward | NADPH | 310000 | 1/min | NaN | 32 | from E. coli, with CoCl2, wild-type enzyme, pH 7.0-8.5, temp >37C (40-60C) | Kuzuyama2000, Br286428 |
| Kcat | Forward | NADPH | 130000 | 1/min | NaN | 32 | from E. coli, with MgCl2, wild-type enzyme, pH 7.0-8.5, temp >37C (40-60C) | Kuzuyama2000, Br286428 |
| Kcat | Forward | NADPH | 590000 | 1/min | NaN | 32 | from E. coli, with MgCl2, wild-type enzyme, pH 7.0-8.5, temp >37C (40-60C) | Kuzuyama2000, Br286428 |
| Parameter | Direction | Substrate | Value | Unit | Weight | Description | Reference | |
|---|---|---|---|---|---|---|---|---|
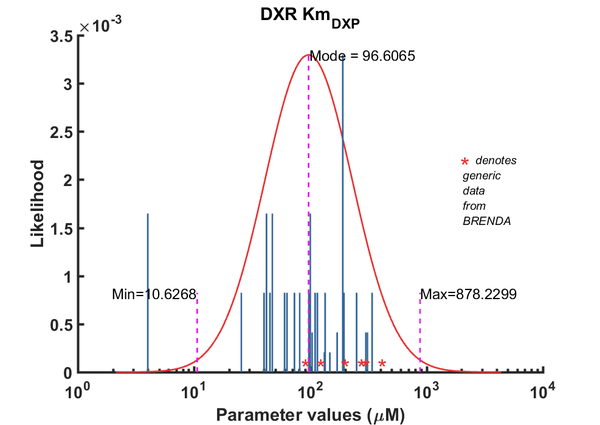
| Km | Forward | DXP | 42 | µM | 64 | from Mycobacterium tuberculosis, transformed into competent E. coli BL21 (DE3) cells. pH7.5, 25C, using three different cofactors Mg2+, Mn2+ and Co2+. And NADPH /NADH BRENDA_lit: 654748, | Argyrou2004, Br654748 | |
| Km | Forward | DXP | 100 | µM | 64 | from Mycobacterium tuberculosis, transformed into competent E. coli BL21 (DE3) cells. pH7.5, 25C, using three different cofactors Mg2+, Mn2+ and Co2+. And NADPH /NADH BRENDA_lit: 654748, | Argyrou2004, Br654748 | |
| Km | Forward | DXP | 4 | µM | 64 | from Mycobacterium tuberculosis, transformed into competent E. coli BL21 (DE3) cells. pH7.5, 25C, using three different cofactors Mg2+, Mn2+ and Co2+. And NADPH /NADH BRENDA_lit: 654748, | Argyrou2004, Br654748 | |
| Km | Forward | DXP | 25.5 | µM | 32 | from Toxoplasma gondii (eukaryote), expressed in E. coli, pH 7.6, 30C | Cai2013, 724543 | |
| Km | Forward | DXP | 190 | µM | 128 | DXPS1; in vitro- S. coelicolor gene expressed in E. coli; pH 7.5, 47C. | Cane2001 | |
| Km | Forward | DXP | 47.1 | µM | 64 | from Mycobacterium tuberculosis, expressed in E. coli, pH 7.9, 30C | Dhiman2005, Br674260 | |
| Km | Forward | DXP | 147.2 | µM | 8 | from Coleus forskohlii (now known as Plectranthus barbatus), expressed in E. coli; pH 8.0, 37C, BRENDA_lit: 676660 | Engpraset2005 | |
| Km | Forward | DXP | 170 | µM | 16 | dxr gene from Synechocystis sp. PCC6803 cloned into E. coli. pH 7.8, 37C, BRENDA_lit:671590 | Fernandes2005, Br671590 | |
| Km | Forward | DXP | 45 | µM | 32 | from E. coli, pH 7.6, 37C, recombinant enzyme | Fox2005, BR672040 | |
| Km | Forward | DXP | 300 | µM | 16 | gene from Zymomonas mobilis, expressed in E. coli, 40C, pH 8.0, | Grolle2000, Br286424 | |
| Km | Forward | DXP | 340 | µM | 32 | Gene MtDXRfrom Mycobacterium tuberculosis, room temperature (20-22C), pH7.5 | Henriksson2006, Br671075 | |
| Km | Forward | DXP | 73 | µM | 32 | gene from E. coli, pH 7.7, 37C, with the presence of 1mM MnCl2 and 3mM MgCl2 | Hoeffler2002, Br655439 | |
| Km | Forward | DXP | 97 | µM | 32 | gene from E. coli, pH 7.7, 37C, with the presence of 1mM MnCl2 and 3mM MgCl2 | Hoeffler2002, Br655439 | |
| Km | Forward | DXP | 103.7 | µM | 16 | gene from Francisella tularensis, expressed in E. coli, pH8, 22C | Jawaid2009, Br713353 | |
| Km | Forward | DXP | 250 | µM | 32 | from E. coli, with MnCl2, wild-type enzyme, pH 7.0-8.5, temp >37C (40-60C) | Kuzuyama2000, Br286428 | |
| Km | Forward | DXP | 99 | µM | 32 | from E. coli, with MgCl2, wild-type enzyme, pH 7.0-8.5, temp >37C (40-60C) | Kuzuyama2000, Br286428 | |
| Km | Forward | DXP | 60 | µM | 32 | from E. coli, with CoCl2, wild-type enzyme, pH 7.0-8.5, temp >37C (40-60C) | Kuzuyama2000, Br286428 | |
| Km | Forward | DXP | 115 | µM | 32 | gene MtDXR from Mycobacterium tuberculosis, expressed in E. coli, pH 7.5, 25C. Experimental result, see Table 1. | Liu2012, BR724346 | |
| Km | Forward | DXP | 132 | µM | 8 | The ispC gene from Arabidopsis thaliana, expressed in E. coli, pH 8.0, 37C | Rohdich2006, Br673562 | |
| Km | Forward | DXP | 40 | µM | 32 | 50C. from Thermotoga maritima. Kinetics were measured at different temperature: 50C and 85C, expressed in E. coli, pH 7.5, 50C. | Takenoya2010, Br712892 | |
| Km | Forward | DXP | 110 | µM | 32 | 85C. from Thermotoga maritima. Kinetics were measured at different temperature: 50C and 85C, expressed in E. coli, pH 7.5, 50C. | Takenoya2010, Br712892 | |
| Km | Forward | DXP | 81 | µM | 32 | from E. coli, pH7.6, 37C | Walker2005 | |
| Km | Forward | DXP | 310 | µM | 16 | from E. coli, ph and temp not mentioned. Reference to method also not mentioned specifically. | Wong2007, Br684434 | |
| Km | Forward | DXP | 195 | µM | 32 | from Synechocystis sp. PCC6803, expressed in E. coli, pH7.5, 37C, with three metal ions, Mn2+, Mg2+, Co2+ | Yin2003, Br654877 | |
| Km | Forward | DXP | 134 | µM | 32 | from Synechocystis sp. PCC6803, expressed in E. coli, pH7.5, 37C, with three metal ions, Mn2+, Mg2+, Co2+ | Yin2003, Br654877 | |
| Km | Forward | DXP | 63 | µM | 32 | from Synechocystis sp. PCC6803, expressed in E. coli, pH7.5, 37C, with three metal ions, Mn2+, Mg2+, Co2+ | Yin2003, Br654877 |
| Parameter | Direction | Substrate | Value | Unit | Weight | Description | Reference |
|---|---|---|---|---|---|---|---|
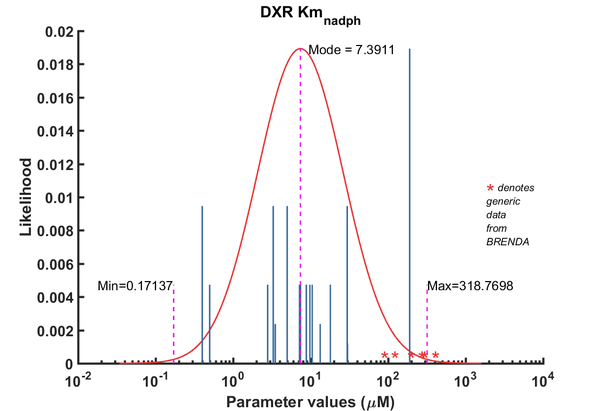
| Km | Forward | NADPH | 5 | µM | 64 | from Mycobacterium tuberculosis, transformed into competent E. coli BL21 (DE3) cells. pH7.5, 25C, using three different cofactors Mg2+, Mn2+ and Co2+. BRENDA_lit: 654748, | Argyrou2004, Br654748 |
| Km | Forward | NADPH | 3.3 | µM | 64 | from Mycobacterium tuberculosis, transformed into competent E. coli BL21 (DE3) cells. pH7.5, 25C, using three different cofactors Mg2+, Mn2+ and Co2+. BRENDA_lit: 654748, | Argyrou2004, Br654748 |
| Km | Forward | NADPH | 0.4 | µM | 64 | from Mycobacterium tuberculosis, transformed into competent E. coli BL21 (DE3) cells. pH7.5, 25C, using three different cofactors Mg2+, Mn2+ and Co2+. BRENDA_lit: 654748, | Argyrou2004, Br654748 |
| Km | Forward | NADPH | 190 | µM | 128 | DXPS1; in vitro- S. coelicolor gene expressed in E. coli; pH 7.5, 47C. | Cane2001 |
| Km | Forward | NADPH | 29.7 | µM | 64 | from Mycobacterium tuberculosis, expressed in E. coli, pH 7.9, 30C | Dhiman2005, Br674260 |
| Km | Forward | NADPH | 3.5 | µM | 16 | dxr gene from Synechocystis sp. PCC6803 cloned into E. coli. pH 7.8, 37C, BRENDA_lit:671590 | Fernandes2005, Br671590 |
| Km | Forward | NADPH | 0.5 | µM | 32 | from E. coli, pH 7.6, 37C, recombinant enzyme | Fox2005, BR672040 |
| Km | Forward | NADPH | 5 | µM | 16 | gene from Zymomonas mobilis, expressed in E. coli, 40C, pH 8.0, | Grolle2000, Br286424 |
| Km | Forward | NADPH | 7.2 | µM | 32 | Gene MtDXRfrom Mycobacterium tuberculosis, room temperature (20-22C), pH7.5 | Henriksson2006, Br671075 |
| Km | Forward | NADPH | 13.3 | µM | 16 | gene from Francisella tularensis, expressed in E. coli, pH8, 22C | Jawaid2009, Br713353 |
| Km | Forward | NADPH | 7.4 | µM | 32 | from E. coli, with MnCl2, wild-type enzyme, pH 7.0-8.5, temp >37C (40-60C) | Kuzuyama2000, Br286428 |
| Km | Forward | NADPH | 18 | µM | 32 | from E. coli, with MgCl2, wild-type enzyme, pH 7.0-8.5, temp >37C (40-60C) | Kuzuyama2000, Br286428 |
| Km | Forward | NADPH | 8.8 | µM | 32 | from E. coli, with CoCl2, wild-type enzyme, pH 7.0-8.5, temp >37C (40-60C) | Kuzuyama2000, Br286428 |
| Km | Forward | NADPH | 9.8 | µM | 32 | gene MtDXR from Mycobacterium tuberculosis, expressed in E. coli, pH 7.5, 25C. Experimental result, see Table 1. | Liu2012, BR724346 |
| Km | Forward | NADPH | 30 | µM | 8 | The ispC gene from Arabidopsis thaliana, expressed in E. coli, pH 8.0, 37C | Rohdich2006, Br673562 |
| Km | Forward | NADPH | 2.8 | µM | 32 | 50C.from Thermotoga maritima. Kinetics were measured at different temperature: 50C and 85C, expressed in E. coli, pH 7.5, 50C. | Takenoya2010, Br712892 |
| Km | Forward | NADPH | 10.5 | µM | 32 | 85C. from Thermotoga maritima. Kinetics were measured at different temperature: 50C and 85C, expressed in E. coli, pH 7.5, 50C. | Takenoya2010, Br712892 |
| Km | Forward | NADPH | 3.3 | µM | 32 | from Synechocystis sp. PCC6803, expressed in E. coli, pH7.5, 37C, with three metal ions, Mn2+, Mg2+, Co2+ | Yin2003, Br654877 |
| Km | Forward | NADPH | 5 | µM | 32 | from Synechocystis sp. PCC6803, expressed in E. coli, pH7.5, 37C, with three metal ions, Mn2+, Mg2+, Co2+ | Yin2003, Br654877 |
| Parameter | Direction | Substrate | Value | Unit | Weight | Description | Reference |
|---|---|---|---|---|---|---|---|
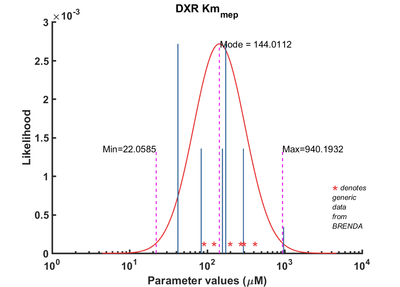
| Km | Reverse | MEP | 42 | µM | 64 | from Mycobacterium tuberculosis, transformed into competent E. coli BL21 (DE3) cells. pH7.5, 25C, using three different cofactors Mg2+, Mn2+ and Co2+. And NADPH /NADH BRENDA_lit: 654748, | Argyrou2004, Br654748 |
| Km | Reverse | MEP | 174 | µM | 64 | from Mycobacterium tuberculosis, expressed in E. coli, pH 7.9, 30C | Dhiman2005, Br674260 |
| Km | Reverse | MEP | 294 | µM | 32 | gene from E. coli, pH 7.7, 37C, with the presence of 1mM MnCl2 and 3mM MgCl2 | Hoeffler2002, Br655439 |
| Km | Reverse | MEP | 158 | µM | 32 | gene from E. coli, pH 7.7, 37C, with the presence of 1mM MnCl2 and 3mM MgCl2 | Hoeffler2002, Br655439 |
| Km | Reverse | MEP | 84 | µM | 32 | gene MtDXR from Mycobacterium tuberculosis, expressed in E. coli, pH 7.5, 25C. Experimental result, see Table 1. | Liu2012, BR724346 |
| Km | Forward | MEP | 972 | µM | 8 | The ispC gene from Arabidopsis thaliana, expressed in E. coli, pH 8.0, 37C | Rohdich2006, Br673562 |
| Parameter | Direction | Substrate | Value | Unit | Weight | Description | Reference | |
|---|---|---|---|---|---|---|---|---|
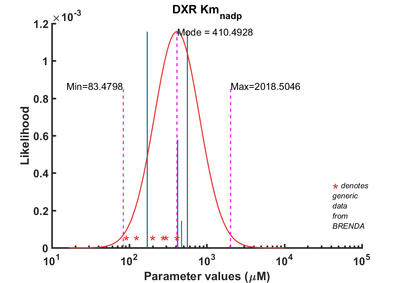
| Km | Reverse | NADP | 170 | µM | 64 | from Mycobacterium tuberculosis, transformed into competent E. coli BL21 (DE3) cells. pH7.5, 25C, using three different cofactors Mg2+, Mn2+ and Co2+. And NADPH /NADH BRENDA_lit: 654748, | Argyrou2004, Br654748 | |
| Km | Reverse | NADP | 560.5 | µM | 64 | from Mycobacterium tuberculosis, expressed in E. coli, pH 7.9, 30C | Dhiman2005, Br674260 | |
| Km | Reverse | NADP | 420 | µM | 32 | gene MtDXR from Mycobacterium tuberculosis, expressed in E. coli, pH 7.5, 25C. Experimental result, see Table 1. | Liu2012, BR724346 | |
| Km | Forward | NADP | 471 | µM | 8 | The ispC gene from Arabidopsis thaliana, expressed in E. coli, pH 8.0, 37C | Rohdich2006, Br673562 |
References
- ↑ Hanekom, A. J. 2006. "Generic kinetic equations for modelling multisubstrate reactions in computational systems biology", MSc Thesis submitted at the University of Stellenbosch
- ↑ Sauro, H.M. "Enzyme kinetics for systems biology"
![V_\mathrm{DXR}= \cfrac{Kcat_\mathrm{forward} \bullet [DXR] \bullet \left(\cfrac{[DXP]}{Km_\mathrm{dxp}}\right) \bullet \left( \cfrac{[NADPH]}{Km_\mathrm{nadph}} \right) \bullet \left( 1 - \cfrac{\cfrac{[DXP]\cdot[NADPH]}{[MEP]\cdot[NADP]}}{Keq}\right)}{\left(1 +\cfrac{[DXP]}{Km_\mathrm{dxp}} + \cfrac{[NADP]}{Km_\mathrm{nadp}}\right) \bullet \left( 1 + \cfrac{[NADPH]}{Km_\mathrm{nadph}} + \cfrac{[MEP]}{Km_\mathrm{mep}}\right)}](/wiki/images/math/0/6/8/068fc5a0c2a7508b25e77f504bb878de.png)