CMK
Contents
The reaction
4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase (CMK) catalyses the production of CDP-ME2P from CDP-ME. This ATP-dependent reaction is the fourth step in the non-mevalonate or methylerythritol-phosphate (MEP) pathway.
Equation Rate
CMK is modelled reversible using random bi-bi generic Michaelis-Menten equation [1]
where :
| Parameter | Description | Units |
|---|---|---|
| VCMK | Reaction rate for CMK | μM/min |
| Kcatforward | Forward turnover number | min-1 |
| KmCDPME | Michaelis-Menten constant for CDPME | μM |
| KmATP | Michaelis-Menten constant for ATP | μM |
| KmCDPME2P | Michaelis-Menten constant for CDPME2P | μM |
| KmADP | Michaelis-Menten constant for ADP | μM |
| Keq | Equilibrium constant | unitless |
| [CDPME] | CDPME concentration | μM |
| [ATP] | ATP concentration | μM |
| [CDPME2P] | CDPME2P concentration | μM |
| [ADP] | ADP concentration | μM |
Parameters
To model CMK using the equation above, a total of 6 parameters are required. For each of this parameter, a lognormal distribution is generated using our PROMENADE pipeline. To include all uncertainties surrounding these parameters, generic parameter values are obtained from BRENDA and is also processed through PROMENADE.
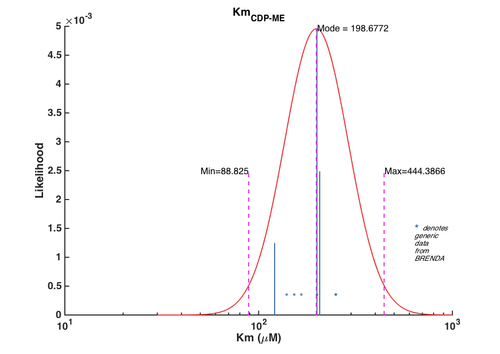
1. Km for CDP-ME
| Parameter value | Error | Weight | Error type | Details | Reference |
|---|---|---|---|---|---|
| 250 | 12.94 | 0.55 | Multiplicative | EC 2.7.1.* Km values from E. coli | BRENDA |
| 153 | 14.55 | 0.22 | Multiplicative | EC 2.7.1.* Km values from all organisms | BRENDA |
| 250 | 13.92 | 0.13 | Multiplicative | EC 2.7.*.* Km values from E. coli | BRENDA |
| 200 | 15.10 | 0.06 | Multiplicative | EC 2.7.*.* Km values from all organisms | BRENDA |
| 140 | 24.91 | 0.03 | Multiplicative | EC 2.*.*.* Km values from E. coli | BRENDA |
| 166 | 19.64 | 0.01 | Multiplicative | EC 2.*.*.* Km values from all organisms | BRENDA |
| 206.70 | 12.50 | 16 | Additive | Wild type M. tuberculosis IspE, transformed into E. coli, pH 7.0, 37C . Values calculated from averaged data from 3 independent experiments. Kinetics for truncated IspE enzymes were also measured for M. tuberculosis, B. pseudomallei in this study but not included in this entry | Eoh, et. al. 2009 [2] |
| 121 | NaN | 8 | Additive | Putative ispE gene from A. aeolicus transformed into E. coli, pH 8.5, 60C | Sgraja, et. al. 2008 [3] |
| 200 | NaN | 32 | Additive | Gene from E. coli, pH not specified, 22C,pH8.0, experiments were done in triplicate. | Tang2011, Br721831 |
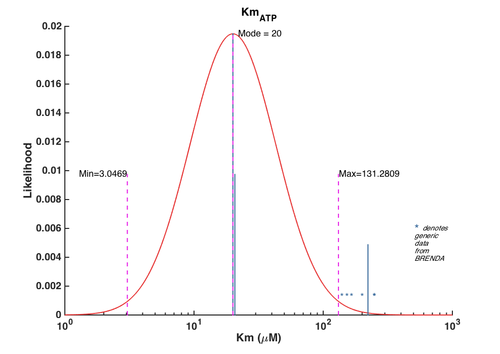
2. Km for ATP
| Parameter value | Error | Weight | Error type | Details | Reference |
|---|---|---|---|---|---|
| 250 | 12.94 | 0.55 | Multiplicative | EC 2.7.1.* Km values from E. coli | BRENDA |
| 153 | 14.55 | 0.22 | Multiplicative | EC 2.7.1.* Km values from all organisms | BRENDA |
| 250 | 13.92 | 0.13 | Multiplicative | EC 2.7.*.* Km values from E. coli | BRENDA |
| 200 | 15.10 | 0.06 | Multiplicative | EC 2.7.*.* Km values from all organisms | BRENDA |
| 140 | 24.91 | 0.03 | Multiplicative | EC 2.*.*.* Km values from E. coli | BRENDA |
| 166 | 19.64 | 0.01 | Multiplicative | EC 2.*.*.* Km values from all organisms | BRENDA |
| 20.70 | 1.20 | 16 | Additive | wild type M. tuberculosis IspE, transformed into E. coli, pH 7.0, 37C . Values calculated from averaged data from 3 independent experiments. Kinetics for truncated IspE enzymes were also measured for M. tuberculosis, B. pseudomallei in this study but not included in this entry | Eoh, et. al. 2009 [4] |
| 222 | 0 | 8 | Additive | Putative ispE gene from A. aeolicus transformed into E. coli, pH 8.5, 60C | Sgraja, et. al. 2008 [5] |
| 20 | 0 | 32 | Additive | Gene from E. coli, pH not specified, 22C,pH8.0, experiments were done in triplicate. | Tang2011, Br721831 |
3. Km for CDP-ME2P
4. Km for ADP
5. Forward Kcat
6. Equilibrium constant (Keq)
Equilibrium constants are calculated from the Van't Hofft Isotherm equation:
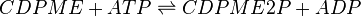
![V_\mathrm{CMK} = \cfrac{\left ( Kcat_\mathrm{forward} \cdot [CMK]\right ) \cdot \left (\cfrac{[CDP-ME]}{Km_\mathrm{CDP-ME}} \right ) \cdot \left(\cfrac{[ATP]}{Km_\mathrm{ATP}} \right) \cdot \left (1 - \cfrac{\cfrac{[CDP-ME2P]\cdot[ADP]}{[CDPME]\cdot[ATP]}}{K_\mathrm{eq}} \right)}{\left(1 + \cfrac{[CDP-ME]}{Km_\mathrm{CDP-ME}} + \cfrac{[ADP]}{Km_\mathrm{ADP}}\right)\cdot \left (1+ \cfrac{[ATP]}{Km_\mathrm{ATP}} +\cfrac{[CDP-ME2P]}{Km_\mathrm{CDP-ME2P}} \right)}](/wiki/images/math/a/2/1/a21c4c09a7ae86ed7025f85bd7f20408.png)