Binding of R2 to A
The ScbR homo-dimer (R2) forms a complex with ScbA (A).
Contents
Chemical equation
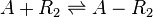
Rate equation
![r= \frac{k^{-}_{3}}{K_{d3}}\cdot [A]\cdot [R_{2}] - k^{-}_{3}\cdot [A-R_{2}]](/wiki/images/math/0/c/d/0cd085b3802a4baeb63922cb1279a5da.png)
Parameters
[NOTE TO MARC: I think the parameter values for the k3 are too arbitrary (see explanation below). Is there any way to choose more reliable ones?]
The parameters of this reaction are the dissociation constant for binding of ScbR to ScbA (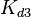 ) and the dissociation rate for binding of ScbR to ScbA (
) and the dissociation rate for binding of ScbR to ScbA ( ). Since no evidence of the existence of the ScbA-ScbR complex has been found so far, our belief is that the interaction between the two proteins should be an unstable/ transient. The values of such complexes according to the literature [1] , lie in the millimolar or micromolar scale.
). Since no evidence of the existence of the ScbA-ScbR complex has been found so far, our belief is that the interaction between the two proteins should be an unstable/ transient. The values of such complexes according to the literature [1] , lie in the millimolar or micromolar scale.
| Parameter | Value | Units | Origin | Remarks |
|---|---|---|---|---|
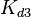
|
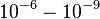 [2] [2]
|

|
Comparison of strong vs weak protein-protein interactions | The range of values correspond to weak protein interactions.  interactions of different proteins were investigated [2] by calculating the interactions of different proteins were investigated [2] by calculating the  by using the Van't Hoff equation by using the Van't Hoff equation 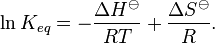
|

|
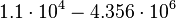 [2] [3] [2] [3]
|

|
From various mechanisms of protein-protein interaction it was deduced that the 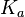 of the reaction should be less than of the reaction should be less than 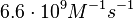 .[3] Therefore the range of .[3] Therefore the range of 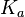 is is 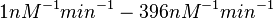 . Therefore the range of the . Therefore the range of the  values is calculated as per values is calculated as per 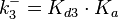 , where the value for , where the value for 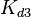 is assumed to be the most plausible is assumed to be the most plausible 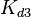 value within the reported range. value within the reported range.
|
Parameters with uncertainty
The probability distributions, adjusted accordingly in order to reflect the above values, are the following:
The most plausible parameter value for the 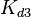 is decided to be
is decided to be 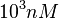 and the confidence interval
and the confidence interval 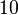 . This means that the mode of the PDF is
. This means that the mode of the PDF is 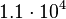 and the range where 95% of the values are found is between
and the range where 95% of the values are found is between 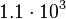 and
and 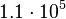 nM.
nM.
In a similar way, the most plausible value for  is
is 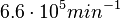 and the confidence interval 6. This means that the mode of the PDF is
and the confidence interval 6. This means that the mode of the PDF is 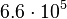 and the range where 95% of the values are found is between
and the range where 95% of the values are found is between 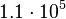 and
and 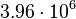
 .
.
The probability distributions for the two parameters, adjusted accordingly in order to reflect the above values, are the following:
The location and scale parameters of the distributions are:
| Parameter | μ | σ |
|---|---|---|
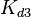
|
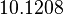
|
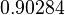
|

|
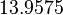
|
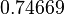
|
References
- ↑ 1.0 1.1 Saliha Ece Acuner Ozbabacan, Hatice Billur Engin, Attila Gursoy, and Ozlem Keskin. Transient protein–protein interactions. Protein Engineering, Design and Selection first published online June 15, 2011
- ↑ 2.0 2.1 2.2 Wesley E. Stites. Protein−Protein Interactions: Interface Structure, Binding Thermodynamics, and Mutational Analysis. Chemical Reviews 1997 97 (5), 1233-1250
- ↑ 3.0 3.1 Janin, Joel. The kinetics of protein-protein recognition. Proteins-Structure Function and Bioinformatics (1997): 153-161.

