Difference between revisions of "Binding of R2 to A"
(→Parameters) |
(→Parameters) |
||
| Line 45: | Line 45: | ||
| <math>630 s^{-1}</math> | | <math>630 s^{-1}</math> | ||
(Range tested: <math>0-10^{3} s^{-1}</math>) | (Range tested: <math>0-10^{3} s^{-1}</math>) | ||
| + | |||
(Bistability range: <math>460-630 s^{-1}</math>) | (Bistability range: <math>460-630 s^{-1}</math>) | ||
|According to Northrup et al. the <math>K_{a}</math> of protein-protein bond formations occur in the order of <math>10^{6} M^{-1}s^{-1}</math>, this value is used to calculate the dissociation rate for the ScbR-ScbA binding. Therefore the range of the <math>k^{-}_{3}</math> values is calculated as per <math>k^{-}_{3}=K_{d3} \cdot K_{a}</math>. | |According to Northrup et al. the <math>K_{a}</math> of protein-protein bond formations occur in the order of <math>10^{6} M^{-1}s^{-1}</math>, this value is used to calculate the dissociation rate for the ScbR-ScbA binding. Therefore the range of the <math>k^{-}_{3}</math> values is calculated as per <math>k^{-}_{3}=K_{d3} \cdot K_{a}</math>. | ||
Revision as of 17:55, 12 October 2015
The ScbR homo-dimer (R2) forms a complex with ScbA (A).
Contents
Chemical equation
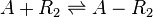
Rate equation
![r= \frac{k^{-}_{3}}{K_{d3}}\cdot [A]\cdot [R_{2}] - k^{-}_{3}\cdot [A-R_{2}]](/wiki/images/math/0/c/d/0cd085b3802a4baeb63922cb1279a5da.png)
Parameters
The parameters of this reaction are the dissociation constant for binding of ScbR to ScbA (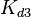 ) and the dissociation rate for binding of ScbR to ScbA (
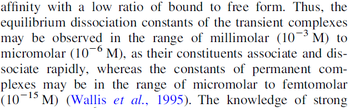
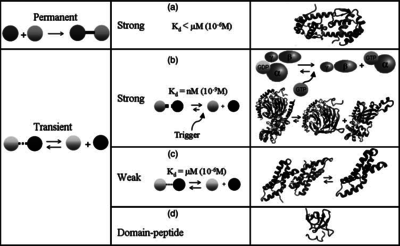
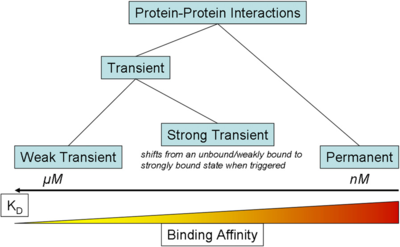
) and the dissociation rate for binding of ScbR to ScbA ( ). Since there is no concrete evidence of the existence of the ScbA-ScbR complex so far, it is possible that the interaction between the two proteins is unstable/ transient and therefore the parameter values reflect this belief. The values of such complexes according to the literature [1] , lie in the millimolar or micromolar scale.
). Since there is no concrete evidence of the existence of the ScbA-ScbR complex so far, it is possible that the interaction between the two proteins is unstable/ transient and therefore the parameter values reflect this belief. The values of such complexes according to the literature [1] , lie in the millimolar or micromolar scale.
| Name | Value | Units | Value in previous GBL model [3] | Remarks-Reference |
|---|---|---|---|---|
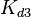
|
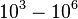 [1] [1]
|

|
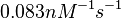
(Range tested: (Bistability range: |
According to the Ozbabacan et al. association constants for transient protein protein interactions lie in the millimolar or micromolar range.
 Ozbabacan et al. 2011[1] |

|
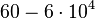 [4] [5] [4] [5]
|

|
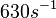
(Range tested: (Bistability range: |
According to Northrup et al. the 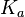 of protein-protein bond formations occur in the order of of protein-protein bond formations occur in the order of 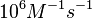 , this value is used to calculate the dissociation rate for the ScbR-ScbA binding. Therefore the range of the , this value is used to calculate the dissociation rate for the ScbR-ScbA binding. Therefore the range of the  values is calculated as per values is calculated as per 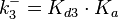 . .
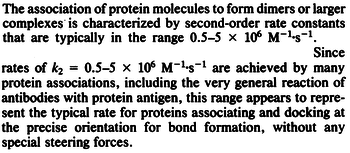 Northrup et al. 1992[5] |
Parameters with uncertainty
Since the values we are using for this parameter correspond to generic association constant values of a wide range of protein-protein interactions and not specifically to GBL or related systems, we wish to explore the whole range of values and investigate the conditions under which the ScbR-ScbA complex formation would be feasible. Therefore, we set the mode of the log-normal distribution for 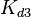 to
to 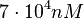 and the confidence interval factor to
and the confidence interval factor to 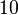 . Thus, the range where 95% of the values are found is between
. Thus, the range where 95% of the values are found is between 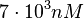 and
and 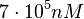 .
.
Similarly, the mode of the log-normal distribution for  is set to
is set to 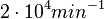 and the confidence interval factor to
and the confidence interval factor to 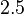 . This means that the range where 95% of the values are found is between
. This means that the range where 95% of the values are found is between 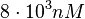 and
and 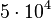
 .
.
The probability distributions for the two parameters, adjusted accordingly in order to reflect the above values, are the following:
The location and scale parameters of the distributions are:
| Parameter | μ | σ |
|---|---|---|
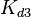
|
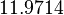
|
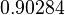
|

|
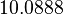
|
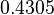
|
References
- ↑ 1.0 1.1 1.2 1.3 Saliha Ece Acuner Ozbabacan, Hatice Billur Engin, Attila Gursoy, and Ozlem Keskin. Transient protein–protein interactions. Protein Engineering, Design and Selection first published online June 15, 2011
- ↑ Perkins J. R., Diboun I., Dessailly B. H., Lees J. G., Orengo C. Transient Protein-Protein Interactions: Structural, Functional, and Network Properties. Structure, 2010;18:10, p. 1233-1243
- ↑ S. Mehra, S. Charaniya, E. Takano, and W.-S. Hu. A bistable gene switch for antibiotic biosynthesis: The butyrolactone regulon in streptomyces coelicolor. PLoS ONE, 3(7), 2008.
- ↑ Janin, Joel. The kinetics of protein-protein recognition. Proteins-Structure Function and Bioinformatics (1997): 153-161.
- ↑ 5.0 5.1 Northrup S.H. and Erickson H.P. Kinetics of protein-protein association explained by Brownian dynamics computer simulation.PNAS 1992;89(8),3338-3342


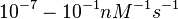 )
)
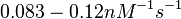 )
)
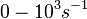 )
)
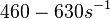 )
)