Difference between revisions of "Basic mevalonate pathway model with limonene synthesis: Construction and Ensemble analysis"
Aliah.hawari (talk | contribs) (→Acknowledgements) |
Aliah.hawari (talk | contribs) (→The simulation) |
||
| Line 7: | Line 7: | ||
This simulation is based on an experiment on the DH10B strain of ''E. coli'' cell cultures that produces limonene that are sequestered into an organic layer. Brief description of the experimental method is as follows: | This simulation is based on an experiment on the DH10B strain of ''E. coli'' cell cultures that produces limonene that are sequestered into an organic layer. Brief description of the experimental method is as follows: | ||
| − | * | + | * The DH10B strain of E. coli was transformed with the limonene production plasmid pJBEI6410. |
| − | * | + | * Three 60 ml cultures (A, B and C), inoculated from individual colonies, were grown in flasks at 30 oC in a shaker incubator. |
| − | * | + | * When cultures reached OD600 of 0.75 they were split into multiple 5 ml aliquots and 1 ml of dodecane overlay was added (timepoint zero). At this point some samples were Induced by the addition of IPTG (25 uM final concentration), other samples were left Uninduced. In addition controls were set-up with limonene added (500 ug/L) to one aliquot of each culture. |
| − | * | + | * All cultures were then incubated at 30 oC in a shaker incubator until the designated timepoints. |
| − | * | + | * At each timepoint (0, 2, 4, 8, 24 hr) samples were removed, OD600 was measured, the dodecane overlay was recovered for limonene quantification, and total RNA was prepared (but no RNAseq was performed on the 24hr samples as RNA quality was too low). The limonene control samples were analysed only at the 8 hr timepoint. The protein fraction of each sample was recovered and stored at -80oC. |
| − | * | + | * The total RNA samples were treated to deplete ribosomal RNA (rRNA) and RNA integrity (RIN values) were determined for each sample prior to sending them for RNAseq |
== Acknowledgements == | == Acknowledgements == | ||
Special thanks to Dr. Christopher Robinson from the SynBioChem Centre University of Manchester for carrying out the experiment and supplying all of the OD readings and limonene titers for this simulation. | Special thanks to Dr. Christopher Robinson from the SynBioChem Centre University of Manchester for carrying out the experiment and supplying all of the OD readings and limonene titers for this simulation. | ||
Revision as of 13:41, 31 January 2018
This model serves as the preliminary MVA pathway framework that we have used as the foundation to debug and design improved MVA models. Although basic, this model was able to provide useful insights into the control of the MVA pathway and suggests strategies for possible engineering. The ensemble analysis carried out for this model have generated a total 8 model generations that have shown a significant improvement in their model scores after each reiteration. A step-by-step ensemble analysis that included scoring the models based on experimental data and identifying influential parameters have found that the proteins that are influential to the pathway are LIMS, MDC, GPPS and IDI. The analysis was also able to shed light on product inhibitions in the pathway that would influence the production of limonene, and also suggests an possible topological engineering design that could accelerate the pathway even more.
The simulation
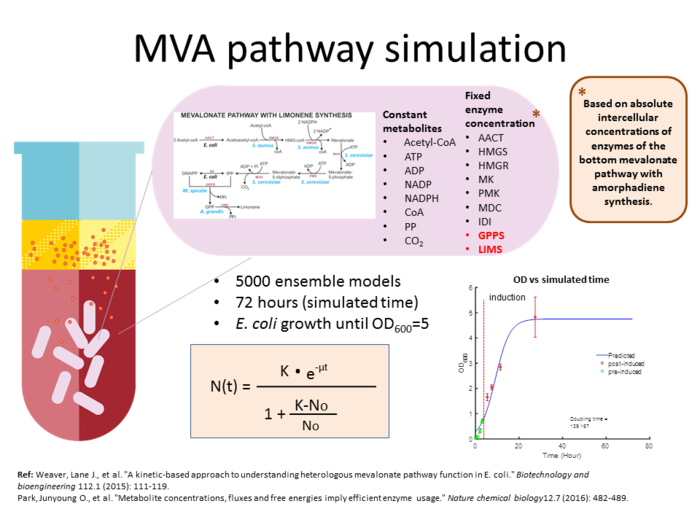
This simulation is based on an experiment on the DH10B strain of E. coli cell cultures that produces limonene that are sequestered into an organic layer. Brief description of the experimental method is as follows:
- The DH10B strain of E. coli was transformed with the limonene production plasmid pJBEI6410.
- Three 60 ml cultures (A, B and C), inoculated from individual colonies, were grown in flasks at 30 oC in a shaker incubator.
- When cultures reached OD600 of 0.75 they were split into multiple 5 ml aliquots and 1 ml of dodecane overlay was added (timepoint zero). At this point some samples were Induced by the addition of IPTG (25 uM final concentration), other samples were left Uninduced. In addition controls were set-up with limonene added (500 ug/L) to one aliquot of each culture.
- All cultures were then incubated at 30 oC in a shaker incubator until the designated timepoints.
- At each timepoint (0, 2, 4, 8, 24 hr) samples were removed, OD600 was measured, the dodecane overlay was recovered for limonene quantification, and total RNA was prepared (but no RNAseq was performed on the 24hr samples as RNA quality was too low). The limonene control samples were analysed only at the 8 hr timepoint. The protein fraction of each sample was recovered and stored at -80oC.
- The total RNA samples were treated to deplete ribosomal RNA (rRNA) and RNA integrity (RIN values) were determined for each sample prior to sending them for RNAseq
Acknowledgements
Special thanks to Dr. Christopher Robinson from the SynBioChem Centre University of Manchester for carrying out the experiment and supplying all of the OD readings and limonene titers for this simulation.