Difference between revisions of "Adenylate kinase"
(→Parameters with uncertainty) |
(→Parameters with uncertainty) |
||
| Line 35: | Line 35: | ||
| The Adenylate kinase was modeled using mass action kinetics with parameters <math>K_1</math> and <math>K_2</math> consistent with the equilibrium constant of the reaction. The equilibrium constant (Keq=2.26) is from Bergmeyer H.U. (1974) page 486<ref name="bergmeyer74">Bergmeyer H.U. (1974) ''Methods of enzymatic analysis'', Publisher: Verlag Chemie (vol 1)</ref>: | | The Adenylate kinase was modeled using mass action kinetics with parameters <math>K_1</math> and <math>K_2</math> consistent with the equilibrium constant of the reaction. The equilibrium constant (Keq=2.26) is from Bergmeyer H.U. (1974) page 486<ref name="bergmeyer74">Bergmeyer H.U. (1974) ''Methods of enzymatic analysis'', Publisher: Verlag Chemie (vol 1)</ref>: | ||
<math>Keq(ATP+AMP \rightarrow 2*ADP, pH=7.4, T=25^oC)=2.26</math><br/> | <math>Keq(ATP+AMP \rightarrow 2*ADP, pH=7.4, T=25^oC)=2.26</math><br/> | ||
| − | In Mass action rate law the relationship is <math>\frac{K_2}{K_1} = K_{eq}</math>. Considering <math>K_{eq} = 2.26</math> we have <math>K_2=2.26</math> and <math>K_1 = 1</math> to be consistent with the equation. The value of <math>K_1</math> and <math>K_2</math> would be varied based on the uncertainty on <math>K_{eq}</math> value mentioned in the following table. | + | In Mass action rate law the relationship is <math>\frac{K_2}{K_1} = K_{eq}</math>. Considering <math>K_{eq} = 2.26</math> we have <math>K_2=2.26</math> and <math>K_1 = 1</math> to be consistent with the equation. The value of <math>K_1</math> and <math>K_2</math> would be varied based on the uncertainty on <math>K_{eq}</math> value mentioned in the following table. The percentage of error in Keq is <math>\approx 4</math>. With similar percent of error the value for <math>K_1 = 1 \pm 0.04</math> and <math>K_2 = 2.26 \pm 0.09044</math> |
|} | |} | ||
Revision as of 16:34, 28 May 2014
Adenylate kinase is a phosphotransferase enzyme that catalyzes the interconversion of adenine nucleotides.
Contents
Chemical equation
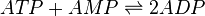
Rate equation
Reversible mass action rate law is used
![K_{1}[ATP][AMP] - K_{2}[ADP]^2](/wiki/images/math/7/f/c/7fc054b0299ef7cf1db75a18a2860588.png)
Parameter values
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
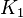
|
1 [1] | HeLa cell line | |
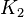
|
2.26 [1] |
Parameters with uncertainty
| Parameter | Value |
|---|---|
Failed to parse (Cannot store math image on filesystem.): K_1=1
, 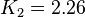
|
The Adenylate kinase was modeled using mass action kinetics with parameters 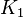 and and 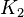 consistent with the equilibrium constant of the reaction. The equilibrium constant (Keq=2.26) is from Bergmeyer H.U. (1974) page 486[2]: consistent with the equilibrium constant of the reaction. The equilibrium constant (Keq=2.26) is from Bergmeyer H.U. (1974) page 486[2]:
|
Equilibrium constant
| Equilibrium constant | Conditions | Source |
|---|---|---|
| 0.48+/-0.015 (mean+/-SEM; n=7) | pH=7, T=25°C, 10mM Mg2+ | NIST database "Thermodynamics of Enzyme-Catalyzed Reactions" entry [61ATK/BUR_640] from Atkinson et al. (1961) [3] Table 2 Therefore, Keq(forward) = 0.48 +/-0.015 (n=7; mean+/-SEM calculated from individual measurements). |
References
- ↑ 1.0 1.1 Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 (doi)
- ↑ Bergmeyer H.U. (1974) Methods of enzymatic analysis, Publisher: Verlag Chemie (vol 1)
- ↑ Atkinson, M. R., Burton, R. M. and Morton, R. K. (1961) Biochem J. 78(4):813–820. (pmid: 13684980)
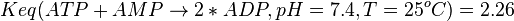
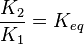 . Considering
. Considering 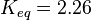 we have
we have 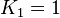 to be consistent with the equation. The value of
to be consistent with the equation. The value of  value mentioned in the following table. The percentage of error in Keq is
value mentioned in the following table. The percentage of error in Keq is 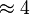 . With similar percent of error the value for
. With similar percent of error the value for 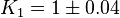 and
and