Difference between revisions of "ATPase"
(→Parameters with uncertainty) |
|||
| Line 25: | Line 25: | ||
==Parameters with uncertainty== | ==Parameters with uncertainty== | ||
| − | * Three value of K for ATPase has been reported in Hernandez (2011) et. al. <ref name="Hernandez2011"></ref>: 0.0042, 0.0045 for Normoxia and 0.003 for Hypoxia. Taking the average and calculating the Std. Dev. the value is <math>0.0039 \pm 0.00079</math> | + | * Three value of K for ATPase has been reported in Hernandez (2011) et. al. <ref name="Hernandez2011"></ref>: 0.0042, 0.0045 for Normoxia and 0.003 for Hypoxia. Taking the average and calculating the Std. Dev. the value is <math>0.0039 \pm 0.00079</math>. Hernandez (2011) et. al. <ref name="Hernandez2011"></ref> reported these values to be adjusted. As the Vmax values were reported in math>U\cdot(\text{mg protein})^{-1}</math>, therefore we adjust these value by multiplying with 65. This gives the value <math>0.2535 \pm 0.05</math> |
{|class="wikitable" | {|class="wikitable" | ||
| Line 34: | Line 34: | ||
|- | |- | ||
|<math>K</math> | |<math>K</math> | ||
| − | |<math>0. | + | |<math>0.2535 \pm 0.05</math> |
|HeLa cell line | |HeLa cell line | ||
| | | | ||
Latest revision as of 15:22, 28 October 2014
ATPases catalyze the decomposition of ATP into ADP and a free phosphate ion. [1]
Contents
Chemical reaction
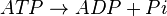
Rate equation
Mass action rate law is used in which 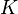 was the rate constant and ATP was the substrate concentration
was the rate constant and ATP was the substrate concentration
![v = K \times [ATP]](/wiki/images/math/d/e/2/de297a3bd10707da7473d1ddc90ec461.png)
Parameter values
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
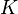
|
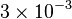 [2] [2]
|
HeLa cell line |
Parameters with uncertainty
- Three value of K for ATPase has been reported in Hernandez (2011) et. al. [2]: 0.0042, 0.0045 for Normoxia and 0.003 for Hypoxia. Taking the average and calculating the Std. Dev. the value is
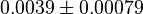 . Hernandez (2011) et. al. [2] reported these values to be adjusted. As the Vmax values were reported in math>U\cdot(\text{mg protein})^{-1}</math>, therefore we adjust these value by multiplying with 65. This gives the value
. Hernandez (2011) et. al. [2] reported these values to be adjusted. As the Vmax values were reported in math>U\cdot(\text{mg protein})^{-1}</math>, therefore we adjust these value by multiplying with 65. This gives the value 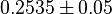
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
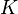
|
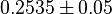
|
HeLa cell line |