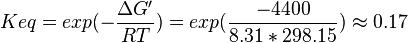3-phosphoglycerate mutase
Phosphoglycerate mutase (PGAM) is an enzyme that catalyzes the internal transfer of a phosphate group from C-3 to C-2 which results in the conversion of 3-phosphoglycerate (3PG) to 2-phosphoglycerate (2PG).
Contents
Chemical equation
Rate equation
Mono-substrate reversible Michaelis-Menten equation is used. [1]
![\frac{V_{mf}\frac{[3PG]}{Km_{3PG}}-V_{mr}\frac{[2PG]}{Km_{2PG}}}{1 + \frac{[3PG]}{Km_{3PG}} + \frac{[2PG]}{Km_{2PG}}}](/wiki/images/math/6/f/e/6fed036845e1a93d67d5db4525e750af.png)
Parameter values
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
|
0.94 [2] | 
|
HeLa cell line | |
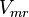
|
0.36 [2] | 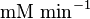
| ||
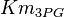
|
0.19[1] | mM | ||
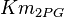
|
0.12[1] | mM |
Parameters with uncertainty
- Two values of
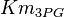 are found for Human cells. They are reported as 0.28[3] and 0.12[1]. Averaging them gives mean value of 0.2 and 0.08
are found for Human cells. They are reported as 0.28[3] and 0.12[1]. Averaging them gives mean value of 0.2 and 0.08
- As the value of the
 does not depend on the organism, the mean and std. dev. of the distribution can be calculated from the various values reported in the literature. [5]
does not depend on the organism, the mean and std. dev. of the distribution can be calculated from the various values reported in the literature. [5] 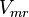 can be sampled based on Haldane equation
can be sampled based on Haldane equation
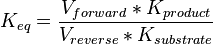 or Alternatively,
or Alternatively,
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
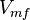
|
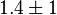 [2] [2]
|
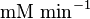
|
HeLa cell line | |
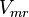
|
Sampled based on Haldane equation 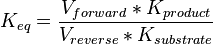
|
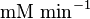
| ||
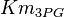
|
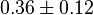
|
mM | Multiple tissue of Human cell | |
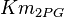
|
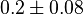
|
mM | Multiple tissue of Human cell |
Equilibrium constant
| Equilibrium constant | Conditions | Source |
|---|---|---|
| 0.15 | pH=7, T=25°C | Voet et al.[6] from Newshole et al. (1973) [7]p 97:
|
| 0.17 | pH=7, T=25°C | Lehninger, (1975)[8] p 412:
|
| 0.167 | pH=7, T=297.15 K | From Meyerhof et al. 1949 (NIST database[9] [49MEY/OES_1388]) |
| 0.19 | pH=7.2, T=25°C | From Chiba et al. 1959 (NIST database[9] [59CHI/SUG_1391]) |
| 0.20 | pH=6, T=25°C | From Grisolia et al. 1975 (NIST database[9] [75GRI/CAR_1396]) |
Averaging all those values gives 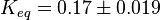
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 (doi)
- ↑ 2.0 2.1 2.2 Marín-Hernández A , Rodríguez-Enríquez S, Vital-González P A, et al. (2006). Determining and understanding the control of glycolysis in fast-growth tumor cells. Flux control by an over-expressed but strongly product-inhibited hexokinase. FEBS J., 273 , pp. 1975–1988(doi)
- ↑ 3.0 3.1 Marin-Hernandez, A., Gallardo-Perez, J. C., Ralph, S. J., Rodriguez-Enriquez, S. & Moreno-Sanchez, R. (2009), HIF-1α modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms. Mini-Rev. Med. Chem. 9, 1084–1101
- ↑ de Atauri, P.; Repiso, A.; Oliva, B.; Vives-Corrons, J.L.; Climent, F.; Carreras, J. (2005), Characterization of the first described mutation of human red blood cell phosphoglycerate mutase, Biochim. Biophys. Acta 1740, 403-410
- ↑ Achcar, F., Kerkhoven, E. J., Bakker, B. M., Barrett, M. P., Breitling, R. (2012), Dynamic modelling under uncertainty: the case of Trypanosoma brucei energy metabolism, PLoS Comput. Biol. 8, e1002352.
- ↑ Voet, D., Voet., J.G. and Pratt, C. W. (1999) Fundamentals of biochemistry, Wiley
- ↑ Newshole, E.A. and Stuart, C. (1973) Regulation in Metabolism, Wiley
- ↑ Lehninger, A.L. (1975) Biochemistry (2nd edn), Worth
- ↑ 9.0 9.1 9.2 Goldberg R.N., Tewari Y.B. and Bhat T.N. (2004) Bioinformatics 20(16):2874-2877 [pmid: 15145806]
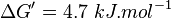 ,
, 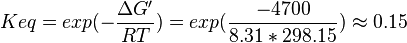
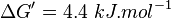 ,
,