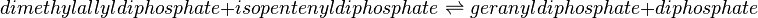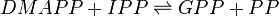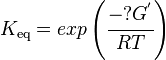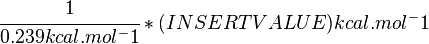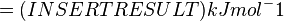Geranyl diphosphate synthase (GPPS)
Revision as of 12:01, 24 March 2016 by Aliah.hawari (talk | contribs) (→Metabolite Background Information)
You can go back to main page of the kinetic model here.
Legend:
| Have not started ·· | 1 -2 data found ·· | 3-4 data found ·· | sufficient data found/estimated ·· | data distribution generated ·· | data sampled |
| to do | DONE! |
Contents
What we know
Issues
Strategies
Reaction catalysed
Metabolite and Enzyme Background Information
Long metabolite names are abbreviated in the model for clarity and standard identification purposes.
| Metabolite | Abbreviation | Chemical Formula | Molar mass (g/mol) | ChEBI | ChEMBL | PubChem | MetaCyc |
|---|---|---|---|---|---|---|---|
| dimethylallyl diphosphate | DMAPP | ||||||
| isopentenyl diphosphate | IPP | ||||||
| geranyl diphosphate | GPP | C10H20O7P2 | 314.209 | 17211 | 41432 | 445995 | |
| diphosphate | PP | O7P2 | 173.94 | 644102 | |||
| geranyl diphosphate synthase | GPPS | CPLX-8656 |
Equation Rate
- Failed to parse (Cannot store math image on filesystem.): V_\mathrm{GPPS} = Vmax_\mathrm{forward} * \cfrac { \left (\cfrac{[DMAPP]}{Km_\mathrm{DMAPP}} * \cfrac{[IPP]}{Km_\mathrm{IPP}}\right )* \left ( 1 - \cfrac {[GPP]*[PP]}{[GPP]*[PP]*K_\mathrm{eq}} \right )}{\left (1 + \cfrac {[IPP]}{Km_\mathrm{IPP}} + \cfrac {[PP]}{Km_\mathrm{PP}} \right ) * \left ( 1 + \cfrac {[DMAPP]}{Km_\mathrm{DMAPP}} + \cfrac {[GPP]}{Km_\mathrm{GPP}} \right )}
| Parameter | Description |
|---|---|
| VGPPS | Reaction rate for Geranyl diphosphate synthase |
| Vmaxforward | Maximum reaction rate towards the production of GPP |
| KmGPP | Michaelis-Menten constant for GPP |
| KmIPP | Michaelis-Menten constant for IPP |
| KmPP | Michaelis-Menten constant for PP |
| KmDMAPP | Michaelis-Menten constant for DMAPP |
| Keq | Equilibrium constant |
| [GPP] | GPP concentration |
| [DMAPP] | DMAPP concentration |
| [IPP] | DMAPP concentration |
| [PP] | PP concentration |
Strategies for estimating the kinetic parameter values
Calculating the Equilibrium Constant
The equilibrium constant can be calculated using the Van't Hoff Isotherm equation:
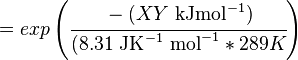
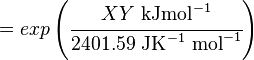
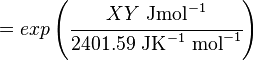
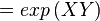
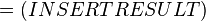
where;
| Keq | Equilibrium constant |
| -?G° | Gibbs free energy change. For (INSERT ENZYME) it is (INSERT VALUE) kJmol-1 |
| R | Gas constant with a value of 8.31 JK-1mol-1 |
| T | Temperature which is always expressed in kelvin |
Standard Gibbs Free energy
Standard Gibbs Free energy for (INSERT ENZYME) from MetaCyc (EC 4.2.3.16) is (INSERT VALUE) kcal/mol [1].
SI derived unit for Gibbs free energy is Joules per mol (J mol-1). 1 kJ·mol-1 is equal to 0.239 kcal·mol-1.
Therefore, the Gibbs free energy for (INSERT ENZYME) in kJ mol-1 is:
Published Kinetic Parameter Values
Km Values
| Km (mM) | Unit | Substrate / Product | Directionality | Organism | References |
|---|---|---|---|---|---|
| Value | unit | substrate | directionality | organism | Ref |
Vmax values
| Vmax | Unit | Directionality | Organism | References |
|---|---|---|---|---|
| Value | µmol/min/mg (unit) | directionality | Organism | References |
Kcat values
| Kcat | Unit | Organism | Reference |
|---|---|---|---|
| value | s-1 | Organism | ref e.g. Alonso 1992 [2] |
Extracting Information from (INSERT SUBSTRATE/PRODUCT) Production Rates
| Amount produced (mg/L) | Time (H) | Organism | Description | Reaction Flux (µM/s) |
|---|---|---|---|---|
| X | X | Y | Z | Z |
| X | X | Y | Z | Z |
| X | X | Y | Z | Z |
| X | X | Y | Z | Z |
| X | X | Y | Z | Z |
| X | X | Y | Z | Z |
Simulations
References
- ↑ Latendresse M. (2013). "Computing Gibbs Free Energy of Compounds and Reactions in MetaCyc."
- ↑ Alonso et. al. 1992. "Purification of 4S-Limonene Synthase, a Monoterpene Cyclase from the Glandular Trichomes of Peppermint (Mentha x piperita) and Spearmint (Mentha spicata)", The Journal of Biological Chemistry, 267(11):7582-7587