Binding of R2 to OR operator
The ScbR homo-dimer (R2) binds to its own gene operator (OR) and represses its mRNA transcription.
Contents
Chemical equation
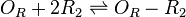
Rate equation
![r= \frac{k^{-}_{1}}{K_{d1}}\cdot [O_{R}]\cdot [R_{2}]^{2} - k^{-}_{1}\cdot [O_{R}-R_{2}]](/wiki/images/math/1/8/d/18df3dce2aa50b661e6a034fc1d8b05f.png)
Parameters
The parameters of this reaction are the dissociation constant for binding of ScbR to OR (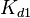 ) and the dissociation rate for binding of ScbR to OR (
) and the dissociation rate for binding of ScbR to OR (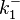 ).
).
| Name | Value | Units | Value in previous GBL models | Remarks-Reference |
|---|---|---|---|---|
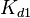
|
 [1] [2] [3] [4] [1] [2] [3] [4]
|

|
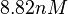
(Range tested: |
TetR-tetO interaction and TetR-like Rv3066 from M. tuberculosis Repressor protein TetR binds to the operator tetO, repressing its own
expression and that of the efflux determinant tetA. Similar structure and activity as ScbR binding to OR |
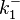
|
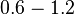 [4] [5] [4] [5]
|

|
N/A | SPR of a TetR-like protein (RolR) on a Gram and GC content ~ 50-60% from Corynebacterium glutamicum |
Parameters with uncertainty
The most plausible parameter value for the 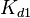 is decided to be
is decided to be 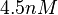 and the confidence interval
and the confidence interval 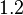 . This means that the mode of the PDF is 4.5 and the range where 95% of the values are found is between 3.75 and 5.4 nM.
. This means that the mode of the PDF is 4.5 and the range where 95% of the values are found is between 3.75 and 5.4 nM.
In a similar way, the most plausible value for 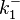 is
is 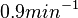 and the confidence interval
and the confidence interval 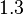 . This means that the mode of the PDF is 0.9 and the range where 95% of the values are found is between 0.6923 and 1.17
. This means that the mode of the PDF is 0.9 and the range where 95% of the values are found is between 0.6923 and 1.17  .
.
The probability distributions for the two parameters, adjusted accordingly in order to reflect the above values, are the following:
The location and scale parameters of the distributions are:
| Parameter | μ | σ |
|---|---|---|
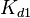
|
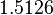
|
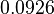
|
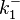
|
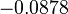
|
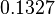
|
References
- ↑ Kamionka A, Bogdanska-Urbaniak J, Scholz O, Hillen W. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Research. 2004;32(2):842-847.
- ↑ Bolla JR, Do SV, Long F, et al. Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis. Nucleic Acids Research. 2012;40(18):9340-9355.
- ↑ Ahn SK, Tahlan K, Yu Z, Nodwell J. Investigation of Transcription Repression and Small-Molecule Responsiveness by TetR-Like Transcription Factors Using a Heterologous Escherichia coli-Based Assay. Journal of Bacteriology. 2007;189(18):6655-6664.
- ↑ 4.0 4.1 Sylwia Kedracka-Krok, Andrzej Gorecki, Piotr Bonarek, and Zygmunt Wasylewski. Kinetic and Thermodynamic Studies of Tet Repressor−Tetracycline Interaction. Biochemistry 2005 44 (3), 1037-1046
- ↑ Li T, Zhao K, Huang Y, et al. The TetR-Type Transcriptional Repressor RolR from Corynebacterium glutamicum Regulates Resorcinol Catabolism by Binding to a Unique Operator, rolO. Applied and Environmental Microbiology. 2012;78(17):6009-6016.


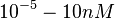 )
)