Binding of R2 to A
The ScbR homo-dimer (R2) forms a complex with ScbA (A).
Contents
Chemical equation
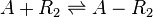
Rate equation
![r= \frac{k^{-}_{3}}{K_{d3}}\cdot [A]\cdot [R_{2}] - k^{-}_{3}\cdot [A-R_{2}]](/wiki/images/math/0/c/d/0cd085b3802a4baeb63922cb1279a5da.png)
Parameters
The parameters of this reaction are the dissociation constant for binding of ScbR to ScbA (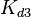 ) and the dissociation rate for binding of ScbR to ScbA (
) and the dissociation rate for binding of ScbR to ScbA ( ). Since no evidence of the existence of the ScbA-ScbR complex has been found so far, our belief is that the interaction between the two proteins should be an unstable/ transient. The values of such complexes according to the literature [1] , lie in the millimolar or micromolar scale.
). Since no evidence of the existence of the ScbA-ScbR complex has been found so far, our belief is that the interaction between the two proteins should be an unstable/ transient. The values of such complexes according to the literature [1] , lie in the millimolar or micromolar scale.
| Parameter | Value | Units | Origin | Remarks |
|---|---|---|---|---|
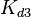
|
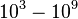 [2] [2]
|

|
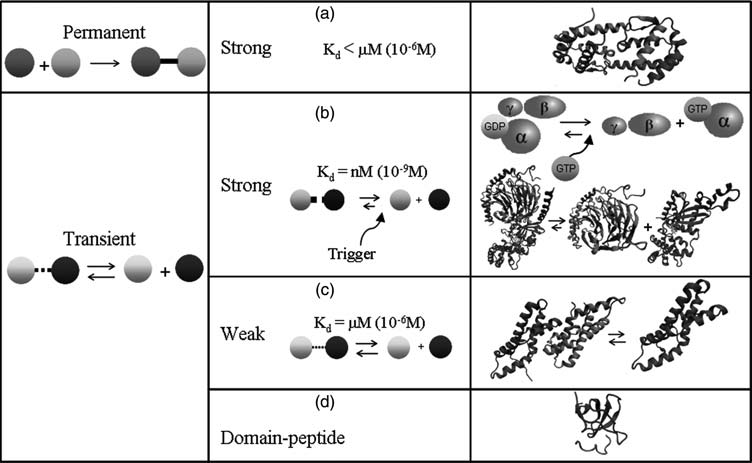
Comparison of strong vs weak protein-protein interactions | Range of values that correspond to weak protein interactions |

|
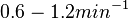 [2] [3] [2] [3]
|

|
Generalistic mechanisms of protein-protein interaction |
Parameters with uncertainty
The most plausible parameter values and the confidence interval were decided to be:
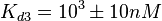
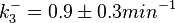
The probability distributions, adjusted accordingly in order to reflect the above values, are the following:
References
- ↑ Saliha Ece Acuner Ozbabacan, Hatice Billur Engin, Attila Gursoy, and Ozlem Keskin. Transient protein–protein interactions. Protein Engineering, Design and Selection first published online June 15, 2011
- ↑ 2.0 2.1 Wesley E. Stites. Protein−Protein Interactions: Interface Structure, Binding Thermodynamics, and Mutational Analysis. Chemical Reviews 1997 97 (5), 1233-1250
- ↑ Janin, Joel. The kinetics of protein-protein recognition. Proteins-Structure Function and Genetics 28.2 (1997): 153-161.