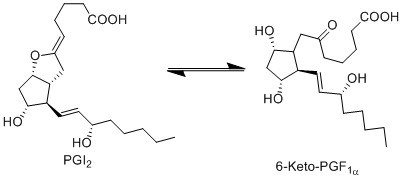
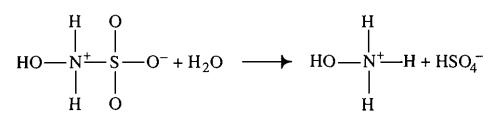
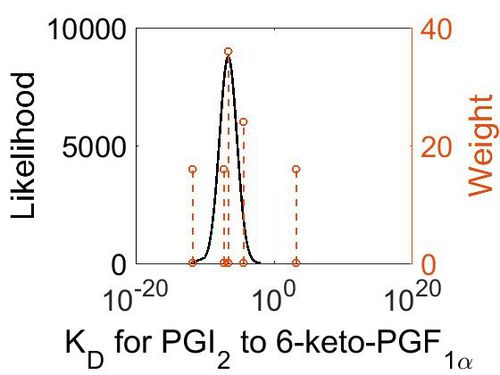
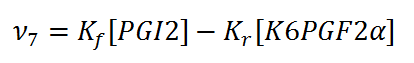Transformation of PGI2 to 6-K-PGF1α
Due to ring strain, PGI2 is rapidly hydrolysed to 6-keto-PGF1a. This is a non-enzymatic reaction and results in the formation of a hydroxyl group at C9 and a ketone at C6, by incorporating two hydrogens and one oxygen.
Contents
Reaction
Chemical equation
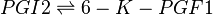
Rate equation
Parameters
Note that the literature values are the same as reaction 6.
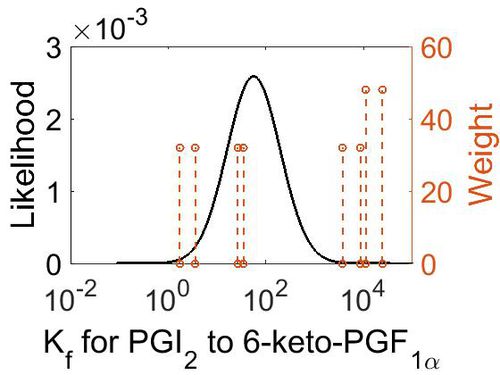
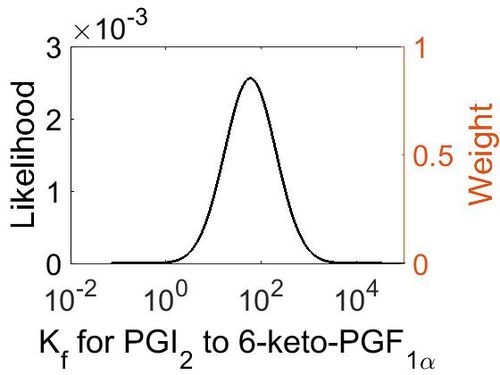
Association Rate Constant (Kf)
| Value | Units | Conditions | Substrate | Weight | Reference |
|---|---|---|---|---|---|
| 3.7e3 ± 0.1e3 | M-1 s-1 | NaCl04 (0.1 - 0.2 M)
Temperature: 25°C |
3 | 32 | [1] |
| 8.7e3 | M-1 s-1 | KCl (1 M)
Temperature: 25°C |
3 | 32 | [1] |
| 1.1e4 ± 0.1e4 | M-1 s-1 | NaCl04 (0.1 - 0.2 M)
Temperature: 25°C |
4 | 48 | [1] |
| 2.4 e4 | M-1 s-1 | KCl (1M)
Temperature: 25°C |
4 | 48 | [1] |
| 3.7e3 ± 0.1e3 | M-1 s-1 | NaCl04 (0.1 - 0.2 M)
Temperature: 25°C |
5 | 32 | [1] |
| 3.6 ± 0.2 | M-1 s-1 | NaCl04 (0.1 - 0.2 M)
Temperature: 25°C |
6 | 32 | [1] |
| 1.7 ± 0.1 | M-1 s-1 | NaCl04 (0.1 - 0.2 M)
Temperature: 25°C |
7 | 32 | [1] |
| 26.7 ± 0.9 | M-1 s-1 | NaCl04 (0.1 - 0.2 M)
Temperature: 25°C |
8 | 32 | [1] |
| 35 | M-1 s-1 | KCl (1M)
Temperature: 25°C |
8 | 32 | [1] |
| 2.25 ± 0.12 | M-1 min-1 | 25°C, in imidazole buffer and also in phosphate buffers, | CO2 to H2CO3 | 32 | [2] |
| Mode (M-1 s-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 3.70E+03 | 3.44E+01 | 1.10E+01 | 1.66E+00 |
Dissociation Rate Constant (Kr)
This is a “Dependent parameter”, meaning that the log-normal distribution for this parameter was calculated using multivariate distributions (this is discussed in detail here). As a result, no confidence interval factor or literature values were cited for this parameter.
| Mode | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 5.28E+01 | 5.63E+00 | 1.29E+00 |
Dissociation Constant
| Value | Units | Conditions | Substrate | Weight | Reference |
|---|---|---|---|---|---|
| 0.000038 | mM | Temperature: 35°C
Vector:Mosquito Note: "In solution, it undergoes rapid hydrolysis to form TXB2, a stable but physiologically inactive compound." - therefore they used stable analogues. |
carbocyclic TXA2 (analogue of TXA2) | 24 | [3] |
| 0.00000023 | mM | Temperature: 35°C
Vector:Rabbit cultured astrocytes Note: "In solution, it undergoes rapid hydrolysis to form TXB2, a stable but physiologically inactive compound." - therefore they used stable analogues. |
[3H]IONO NT-126, a TXA z antagonist, | 36 | [4] |
| 1500 ± 500 (excluded) | mM | Temperature: 25°C
In vitro |
Hydroxysulfamic acid | 16 | [5] |
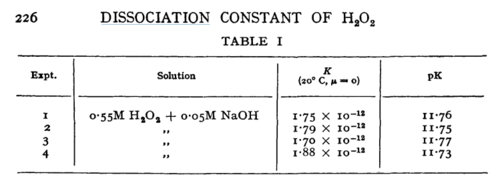
| N/A | Temperature: 20°C
In vitro |
H2O2 | 16 | [5] | |
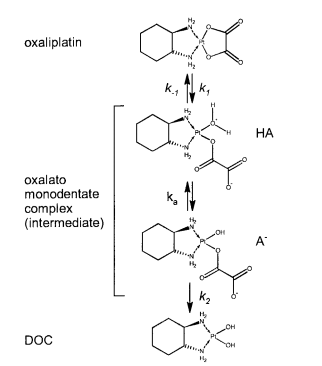
| 5.9 E-8 | N/A | Oxaliplatin | 16 | [6] |
| Mode (M-1 s-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 2.35E-07 | 1.76E+04 | -6.86E+00 | 2.90E+00 |
Related Reactions
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 A. Ross "Vinyl epoxide hydrolysis reactions" J. Am. Chem. Soc., 1982, 104 (6), pp 1658–1665
- ↑ B. Gibbons "Rate of Hydration of Carbon Dioxide and Dehydration of Carbonic Acid at 25" J Biol Chem. 1963 Oct;238:3502-7
- ↑ P. H. Alvarenga "The Function and Three-Dimensional Structure of a Thromboxane A2/Cysteinyl Leukotriene-Binding Protein from the Saliva of a Mosquito Vector of the Malaria Parasite" PLoS Biol. 2010 Nov 30;8(11):e1000547. doi: 10.1371/journal.pbio.1000547.
- ↑ N Nakahata et al. "The Presence of Thromboxane A2 Receptors in Cultured Astrocytes From Rabbit Brain" Brain Res 583 (1-2), 100-104. 1992 Jun 26
- ↑ 5.0 5.1 D. LITTLEJOHN "The dissociation constant and acid hydrolysis rate of hydroxysulfamic acid" Can. J. Chem. 67, 1596 (1989). Cite error: Invalid
<ref>tag; name "Littlejohn1989.E2.80.9D" defined multiple times with different content - ↑ Elin Jerremalm "Hydrolysis of Oxaliplatin—Evaluation of the Acid Dissociation Constant for the Oxalato Monodentate Complex" Journal of Pharmaceutical Sciences Volume 92, Issue 2, February 2003, Pages 436–438