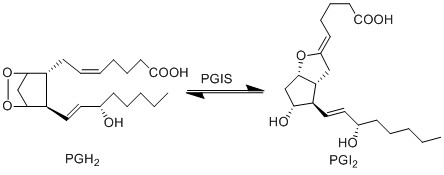
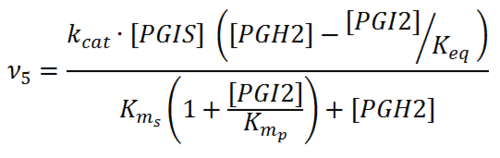Transformation of PGH2 to PGI2
The isomerisation of PGH2 to prostacyclin (PGI2), is performed by prostaglandin I synthase (PGIS). This protein is a member of the CYP P450 family, but unlike most CYP P450 enzymes it does not oxidise PGH2. PGI2 is generated by the rearrangement of the peroxide functional group, whereby a hydroxyl group is formed at C11, and a new epoxide ring is formed between C9 and C6. PGI2 is generated by the sequential action of COX and PGIS, which co-localise in the ER, plasma membrane and nuclear membrane [1].
Contents
Reaction
Chemical equation
Rate equation
Parameters
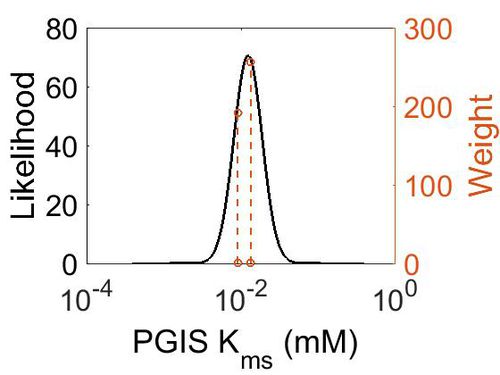
Kms
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 1.33E-02 ± 1.40E-03 | 
|
Human | Expression Vector: Bovine Endothelial and Aorta Cells
Enzyme: Human PGIS pH:7.4 Temperature: 23 |
256 | [2] |
| 9.00E-03 ± 5.00E-03 | 
|
Bovine | Expression Vector: Bovine Endothelial and Aorta Cells
Enzyme: Bovine PGIS pH:7.4 Temperature: 24 |
192 | [3] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.24E-02 | 2.46E+00 | -4.22E+00 | 4.20E-01 |
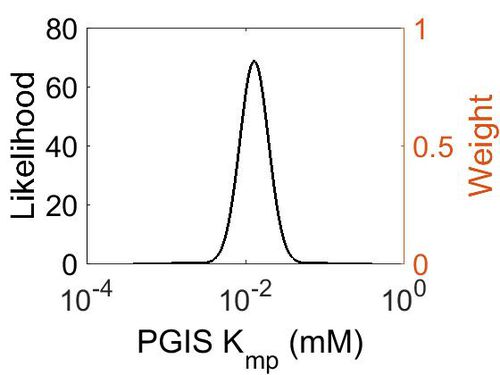
Kmp
| Mode (mM) | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 1.29E-02 | -4.182022605 | 0.414183521 |
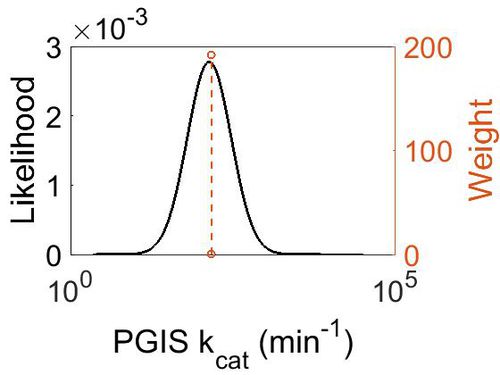
kcat
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 147 ± 45 | per minute | Cattle | Expression Vector: E. Coli
Enzyme: Bovine PGIS pH:7.4 Temperature: 24 |
192 | [4] |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.41E+02 | 1.35E+00 | 5.028213628 | 0.287348692 |
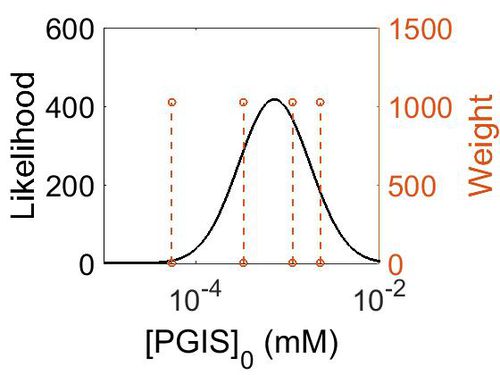
Enzyme concentration
To convert the enzyme concentration from ppm to mM, the following equation was used.
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 412 | 
|
Human | Expression Vector: Urinary bladder
Enzyme: PGIS pH: 7.5 Temperature: 37 °C |
1024 | [5] |
| 206 | 
|
Human | Expression Vector: Lung
Enzyme: PGIS pH: 7.5 Temperature: 37 °C |
1024 | [5] |
| 60.1 | 
|
Human | Expression Vector: Esophagus
Enzyme: PGIS pH: 7.5 Temperature: 37 °C |
1024 | [5] |
| 9.93 | 
|
Human | Expression Vector: Oral Cavity
Enzyme: PGIS pH: 7.5 Temperature: 37 °C |
1024 | [6] |
| Mode (ppm) | Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|---|
| 1.13E+02 | 6.25E-04 | 4.13E+00 | 5.67E+00 | 9.68E-01 |
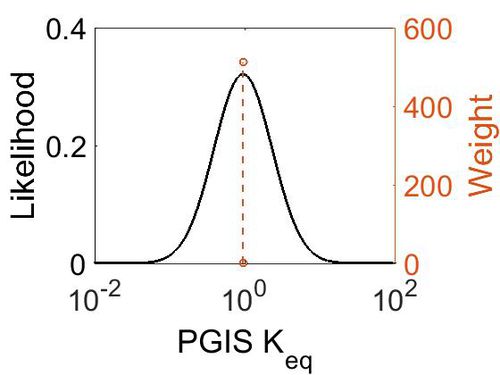
Keq
| Gibbs Free Energy Change | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 0.04 | kcal/mol | Not stated | Estimated
Enzyme: Transacylase Substrate: Product: pH: 7.3 ionic strength: 0.25 |
64 | [7] |
| Mode | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 9.35E-01 | 1.00E+01 | 7.30E-01 | 8.90E-01 |
References
- ↑ Smith, W. L. DeWitt, D. L. Allen, M. L. , Bimodal distribution of the prostaglandin I2 synthase antigen in smooth muscle cells, J Biol Chem (1983), 258, 5922-6.
- ↑ H. C. Yeh, P. Y. Hsu, Characterization of heme environment and mechanism of peroxide bond cleavage in human prostacyclin synthase. Biochim Biophys Acta. 2005 Dec 30;1738(1-3):121-32.
- ↑ Hara S., Isolation and molecular cloning of prostacyclin synthase from bovine endothelial cells. J Biol Chem. 1994 Aug 5;269(31):19897-903.
- ↑ [www.ncbi.nlm.nih.gov/pubmed/8051072 Hara S. , Isolation and molecular cloning of prostacyclin synthase from bovine endothelial cells. J Biol Chem. 1994 Aug 5;269(31):19897-903.]
- ↑ 5.0 5.1 5.2 M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587
- ↑ Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471