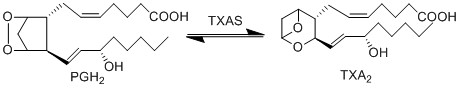
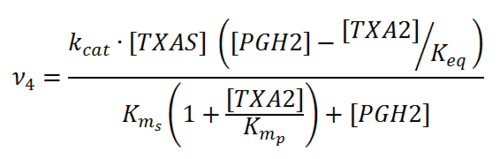Transformation of PGH2 to TXA2
The isomerisation of PGH2 to TXA2 is performed by TXAS. This reaction includes the rearrangement of the peroxide functional group by the protein’s heme group, whereby one oxygen is incorporated into the cyclopentane ring between C11 and C12 to form an oxane ring, whilst the other forms an epoxide group across the oxane ring between C9 and C11. This enzyme is located on the ER membrane and is a member of the cytochrome P450 family based upon the genetic sequence.
TXA2 and TXB2 bind to the TP receptor and have been found to increase T-cell proliferation (Thomas, Rocha et al. 2003). However this species is found in low concentration in the cutaneous compartment, therefore it has been deduced that the species originates from infiltrating cells from the vascular system such as leukocytes and endothelial cells (Sugimoto, Arai et al. 2006).
Contents
Reaction
Chemical equation
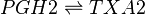
Rate equation
Parameters
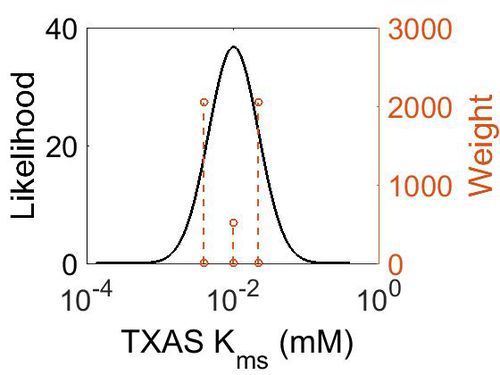
Kms
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 4.00E-03 | 
|
Human | Expression Vector: Human Monocyte
Enzyme: Platelet Free Human Monocyte pH: 7.4 Temperature: 37 |
[1] |
| 1.00E-02 | 
|
Human | Expression Vector: Human mirosome
Enzyme: Human Thromboxane Synthase pH:7.4 Temperature: |
[2] |
| 2.20E-02 | 
|
Human | Expression Vector:Human platelet
Enzyme: Human thromboxane synthase pH: 7.4 Temperature: 37 |
[3] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 9.96E-03 | 2.25E+00 | -3.9721 | 0.78374 |
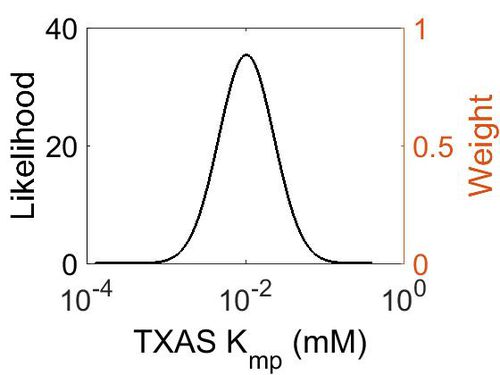
Kmp
| Mode (mM) | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 1.03E-02 | -3.942048511 | 0.797944323 |
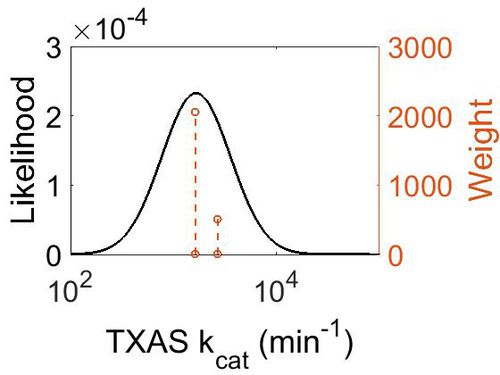
kcat
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 2687 | per minute | Human | Expression Vector: Human Platelets
Enzyme: TXAS pH: 7.4 Temperature:30 |
[4] |
| 1628 | per minute | Human | Expression Vector:Platelet Microsomes
Enzyme: TXAS pH:7.4 Temperature:37 |
[5] |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.67E+03 | 1.25E+00 | 7.467597712 | 0.216816683 |
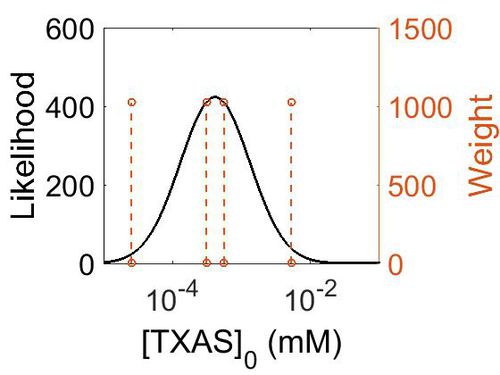
Enzyme concentration
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 949 | 
|
Human | Expression Vector: Platelet
Enzyme: TXAS pH: 7.5 Temperature: 37 °C |
[6] |
| 56.2 | 
|
Human | Expression Vector: Lung
Enzyme: TXAS pH: 7.5 Temperature: 37 °C |
[6] |
| 4.58 | 
|
Human | Expression Vector: Urinary Bladder
Enzyme: TXAS pH: 7.5 Temperature: 37 °C |
[6] |
| 101 | 
|
Human | Expression Vector: Oral Cavity
Enzyme: TXAS pH: 7.5 Temperature: 37 °C |
[7] |
| Mode | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 7.50E+01 | 6.69E+00 | 5.66E+00 | 1.16E+00 |
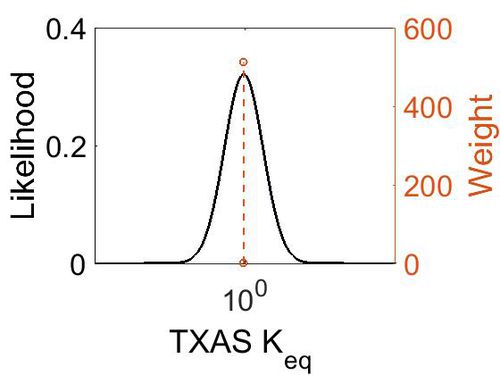
Keq
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 41.08 | kcal/mol | Not stated | Estimated
Enzyme: TXAS Substrate: Arachidonate Product: TXA2 pH: 7.3 ionic strength: 0.25 |
[8] |
| Mode | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 9.35E-01 | 1.00E+01 | 7.30E-01 | 8.90E-01 |
References
- ↑ M. Orlandi, G. Bartolini, Thromboxane A2 synthase activity in platelet free human monocytes. Biochim Biophys Acta. 1994 Dec 8;1215(3):285-90.
- ↑ R. Nusing, S. Schneider-Voss and V. Ullrich, Immunoaffinity purification of human thromboxane synthase. Arch Biochem Biophys. 1990 Aug 1;280(2):325-30.
- ↑ Hecker M. On the mechanism of prostacyclin and thromboxane A2 biosynthesis.J Biol Chem. 1989 Jan 5;264(1):141-50.
- ↑ [www.ncbi.nlm.nih.gov/pubmed/3579292 Hecker M. “Products, kinetics, and substrate specificity of homogeneous thromboxane synthase from human platelets: Development of a novel enzyme assay ” Arch. Biochem. Biophys. 1987, 254, 124-135]
- ↑ [www.ncbi.nlm.nih.gov/pubmed/2999104 Haurand M. “Isolation and characterization of thromboxane synthase from human platelets as a cytochrome P-450 enzyme” J Biol Chem. 1985, 260 (5), 15059-15067]
- ↑ 6.0 6.1 6.2 M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587
- ↑ Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471