Difference between revisions of "Dehydrogenase"
| Line 26: | Line 26: | ||
|1<ref name="Hernandez2011"></ref> | |1<ref name="Hernandez2011"></ref> | ||
|} | |} | ||
| + | |||
| + | |||
| + | ==Parameters with uncertainty== | ||
| + | * This is a pseudo-reaction, modelled using mass action kinetics (i.e., as non-saturable, non-enzymatic reactions) to maintain maximal compatibility with Pyridine Nucleotides balances. The parameter values were adjusted through model simulations and the values chosen were those that best predicted NAD+ concentration (determined experimentally) in Hernandez ''et. al.'' <ref name="Hernandez2011"></ref>. No information is available about the uncertainty of these parameters. As these parameters are strictly positive, they are sampled using a log-normal distribution as are and values. The means are set to the value using fitted by Hernandez et al. <ref name="Hernandez2011"></ref> for the fixed-parameter model. The sampling of the parameters are made to fall within <math>[0.01\times mean \text{ } 1000 \times mean ]</math> to allow a large exploration of the parameter space. | ||
| + | |||
| + | |||
| + | {|class="wikitable" | ||
| + | ! Parameter | ||
| + | ! Value | ||
| + | ! Organism | ||
| + | ! Remarks | ||
| + | |- | ||
| + | |<math>K_{1}</math> | ||
| + | |<math>250 \pm 130078.125</math> | ||
| + | |rowspan="2"|HeLa cell line | ||
| + | |rowspan="2"| | ||
| + | |- | ||
| + | |<math>K_{2}</math> | ||
| + | |1<ref name="Hernandez2011"></ref> | ||
| + | |} | ||
| + | |||
| + | |||
==References== | ==References== | ||
<references/> | <references/> | ||
Revision as of 10:03, 15 May 2014
A dehydrogenase is an enzyme that oxidizes a substrate by a reduction reaction that transfers one or more hydrides (H−) to an electron acceptor, usually Nicotinamide adenine dinucleotide NAD+/NADP.
Contents
Chemical reaction
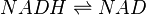
Rate equation
Reversible mass action rate law is used
![K_{1}[NADH] - K_{2}[NAD]](/wiki/images/math/5/7/f/57ff1f0db057e5957f43bd3cf4f50fd1.png)
Parameters
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
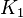
|
250 [1] | HeLa cell line | |
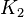
|
1[1] |
Parameters with uncertainty
- This is a pseudo-reaction, modelled using mass action kinetics (i.e., as non-saturable, non-enzymatic reactions) to maintain maximal compatibility with Pyridine Nucleotides balances. The parameter values were adjusted through model simulations and the values chosen were those that best predicted NAD+ concentration (determined experimentally) in Hernandez et. al. [1]. No information is available about the uncertainty of these parameters. As these parameters are strictly positive, they are sampled using a log-normal distribution as are and values. The means are set to the value using fitted by Hernandez et al. [1] for the fixed-parameter model. The sampling of the parameters are made to fall within Failed to parse (Cannot store math image on filesystem.): [0.01\times mean \text{ } 1000 \times mean ] to allow a large exploration of the parameter space.
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
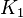
|
Failed to parse (Cannot store math image on filesystem.): 250 \pm 130078.125 | HeLa cell line | |
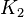
|
1[1] |