Difference between revisions of "Glyceraldehyde-3-phosphate dehydrogenase"
(→Parameters with uncertainty) |
(→Parameters with uncertainty) |
||
| Line 50: | Line 50: | ||
==Parameters with uncertainty== | ==Parameters with uncertainty== | ||
| − | * 4 values of <math>Km_{NADH}</math> have been collected from Lambeir ''et. al.'' (1991)<ref name="Lambeir_1991">Lambeir, A.M.; Loiseau, A.M.; Kuntz, D.A.; Vellieux, F.M.; Michels, P.A.M.; Opperdoes, F.R. (1991), ''The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic properties and comparison with homologous enzymes'', Eur. J. Biochem. 198, 429-435</ref> for different organisms. <math>Km_{NADH} = 0.007</math> for Trypanosoma brucei, <math>Km_{NADH} = 0.01</math> for Homo sapiens, <math>Km_{NADH} = 0.012</math> for Geobacillus stearothermophilus and <math>Km_{NADH} = 0.012</math> for Oryctolagus cuniculus. | + | * 4 values of <math>Km_{NADH}</math> have been collected from Lambeir ''et. al.'' (1991)<ref name="Lambeir_1991">Lambeir, A.M.; Loiseau, A.M.; Kuntz, D.A.; Vellieux, F.M.; Michels, P.A.M.; Opperdoes, F.R. (1991), ''The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic properties and comparison with homologous enzymes'', Eur. J. Biochem. 198, 429-435</ref> for different organisms. <math>Km_{NADH} = 0.007</math> for Trypanosoma brucei, <math>Km_{NADH} = 0.01</math> for Homo sapiens, <math>Km_{NADH} = 0.012</math> for Geobacillus stearothermophilus and <math>Km_{NADH} = 0.012</math> for Oryctolagus cuniculus. Calculating mean and std. dev. from these 4 values gives <math>Km_{NADH} = 0.01025 \pm 0.004063 </math> |
*<ref name = "Ryzlak_1998">Ryzlak, M.T.; Pietruszko, R. (1998), ''Heterogeneity of glyceraldehyde-3-phosphate dehydrogenase from human brain'', Biochim. Biophys. Acta 954, 309-324</ref> | *<ref name = "Ryzlak_1998">Ryzlak, M.T.; Pietruszko, R. (1998), ''Heterogeneity of glyceraldehyde-3-phosphate dehydrogenase from human brain'', Biochim. Biophys. Acta 954, 309-324</ref> | ||
| Line 83: | Line 83: | ||
|- | |- | ||
|<math>Km_{NADH}</math> | |<math>Km_{NADH}</math> | ||
| − | |0. | + | |<math>0.01025 \pm 0.004063</math> |
|mM | |mM | ||
|- | |- | ||
Revision as of 11:06, 29 April 2014
The Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) enzyme serves two functions in glycolytic pathway. First the enzyme transfers a hydrogen (H-) from glyceraldehyde phosphate (Gly3P) to the oxidizing agent Nicotinamide Adenine Dinucleotide (NAD+) to form NADH. Next it adds a phosphate (P) from the cytosol to the oxidized Gly3P to form 1, 3-bisphosphoglycerate.
Contents
Chemical equation
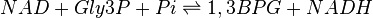
Rate equation
Ordererd Ter-Bi reversible Michaelis-Menten equation for non-interacting substrates. [1]
![v = \frac{ V_{mf}\frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} - V_{mr} \frac{[1,3BPG][NADH]}{K_{1,3BPG}K_{NADH}} }{1 + \frac{[NAD]}{K_{NAD}} + \frac{[NAD][Gly3P]}{K_{NAD}K_{Gly3P}} + \frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} + \frac{[1,3BPG][NADH]}{K_{1,3BPG}K_{NADH}} + +\frac{[NADH]}{K_{NADH}} }](/wiki/images/math/c/e/c/cec09052887411fec63867405d65296f.png)
Parameters
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
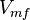
|
0.58 [2] | 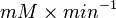
|
HeLa cell line | |
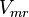
|
0.72[2] | 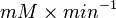
| ||
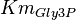
|
0.19[1] | mM | ||
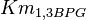
|
0.022[1] | mM | ||
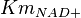
|
0.09[1] | mM | ||
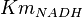
|
0.01[1] | mM | ||
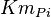
|
29[1] | mM |
Parameters with uncertainty
- 4 values of
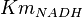 have been collected from Lambeir et. al. (1991)[3] for different organisms.
have been collected from Lambeir et. al. (1991)[3] for different organisms. 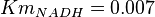 for Trypanosoma brucei,
for Trypanosoma brucei, 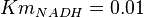 for Homo sapiens,
for Homo sapiens, 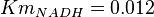 for Geobacillus stearothermophilus and
for Geobacillus stearothermophilus and 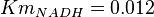 for Oryctolagus cuniculus. Calculating mean and std. dev. from these 4 values gives Failed to parse (Cannot store math image on filesystem.): Km_{NADH} = 0.01025 \pm 0.004063
for Oryctolagus cuniculus. Calculating mean and std. dev. from these 4 values gives Failed to parse (Cannot store math image on filesystem.): Km_{NADH} = 0.01025 \pm 0.004063
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
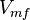
|
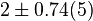 [2] [2]
|
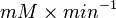
|
HeLa cell line | |
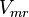
|
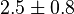 (5)[2] (5)[2]
|
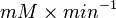
| ||
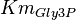
|
0.19[1] | mM | ||
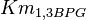
|
0.022[1] | mM | ||
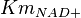
|
0.09[1] | mM | ||
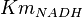
|
Failed to parse (Cannot store math image on filesystem.): 0.01025 \pm 0.004063 | mM | ||
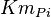
|
29[1] | mM |
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 (doi)
- ↑ 2.0 2.1 2.2 2.3 Marín-Hernández A , Rodríguez-Enríquez S, Vital-González P A, et al. (2006). Determining and understanding the control of glycolysis in fast-growth tumor cells. Flux control by an over-expressed but strongly product-inhibited hexokinase. FEBS J., 273 , pp. 1975–1988(doi)
- ↑ Lambeir, A.M.; Loiseau, A.M.; Kuntz, D.A.; Vellieux, F.M.; Michels, P.A.M.; Opperdoes, F.R. (1991), The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic properties and comparison with homologous enzymes, Eur. J. Biochem. 198, 429-435
- ↑ Ryzlak, M.T.; Pietruszko, R. (1998), Heterogeneity of glyceraldehyde-3-phosphate dehydrogenase from human brain, Biochim. Biophys. Acta 954, 309-324