Difference between revisions of "Glycogen phosphorylase"
(→Rate equation) |
(→Rate equation) |
||
| Line 12: | Line 12: | ||
An alternative rate equation without considering the allosteric regulation is given as <ref name="Lambeth_2002> Lambeth M.J. & Kushmerick M.J. (2002). ''A computational model for glycogenolysis in skeletal muscle''. Ann Biomed Eng 30, 808–827</ref> | An alternative rate equation without considering the allosteric regulation is given as <ref name="Lambeth_2002> Lambeth M.J. & Kushmerick M.J. (2002). ''A computational model for glycogenolysis in skeletal muscle''. Ann Biomed Eng 30, 808–827</ref> | ||
| − | <center><math> \frac{V_{maxf} \frac{Glycogen_{n+1} \times Pi}{K_{iGlyf} \times K_{Pi}} -\frac{V_{maxf} \times K_{Glyb} \times K_{iGlc1P}}{K_{iGlyf} \times K_{Pi} \times Keq} \frac{Glycogen_n \times Glc1P}{K_{Glyb} \times K_{iGlc1P}} }{1} </math></center> | + | <center><math> \frac{V_{maxf} \frac{Glycogen_{n+1} \times Pi}{K_{iGlyf} \times K_{Pi}} -\frac{V_{maxf} \times K_{Glyb} \times K_{iGlc1P}}{K_{iGlyf} \times K_{Pi} \times Keq} \times \frac{Glycogen_n \times Glc1P}{K_{Glyb} \times K_{iGlc1P}} }{1 + \frac{Glycogen_{n+1}}{K_{iGlyf}} + \frac{Pi}{K_{iPi}} + \frac{Glycogen_n}{K_{iGlyb}} + \frac{Glc1P}{K_{iGlc1P}} \frac{Glycogen_{n+1} \times Pi}{K_{Glyf} \times K_{iPi}} + \frac{Glycogen_n \times Glc1P}{K_{Glyb} \times K_{iGlc1P}} } </math></center> |
==Parameter values== | ==Parameter values== | ||
Revision as of 11:51, 1 April 2014
Glycogen phosphorylase (GP) is a dimeric enzyme that catalyses the reaction in which the terminal glucose residue from a glycogen chain is phosphorylated and cleaved from the chain, releasing it as Glc1P.
Chemical equation
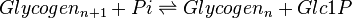
Rate equation
MWC model was used to formualte the rate law. [1]
![\frac{V_{max} \times n \times \frac{[Pi]}{K_{r,Pi}} \left( 1 + \frac{[Pi]}{K_{r,Pi}} + \frac{[Glc1P]}{K_{r,Glc1P}} \right)^{n-1} }{\left( 1 + \frac{[Pi]}{K_{r,Pi}} + \frac{[Glc1P]}{K_{r,Glc1P}} \right)^n + L_u \left( 1 + \frac{[Pi]}{K_{u,Pi}} + \frac{[Glc1P]}{K_{u,Glc1P}} \right)^n \left( \frac{1 + \frac{[Glc6P]}{K_{u,Glc6P}}}{1 + \frac{[AMP]}{K_{r,AMP}} + \frac{[Glc6P]}{K_{r,Glc6P}}} \right)^n } + \frac{V_{max} \times n \times \frac{[Pi]}{K_{r,Pi}} \left( 1 + \frac{[Pi]}{K_{r,Pi}} + \frac{[Glc1P]}{K_{r,Glc1P}} \right)^{n-1} }{\left( 1 + \frac{[Pi]}{K_{r,Pi}} + \frac{[Glc1P]}{K_{r,Glc1P}} \right)^n + L_t \left( 1 + \frac{[Pi]}{K_{t,Pi}} + \frac{[Glc1P]}{K_{t,Glc1P}} \right)^n \left( \frac{1 + \frac{[AMP]}{K_{t,AMP}} \frac{[Glc6P]}{K_{t,Glc6P}}}{1 + \frac{[AMP]}{K_{r,AMP}} + \frac{[Glc6P]}{K_{r,Glc6P}}} \right)^n \left( 1 + \frac{[ATP]}{K_{t,ATP}} \right)^n }](/wiki/images/math/7/e/5/7e599d4304d1f49ee3e3d2d32f556863.png)
An alternative rate equation without considering the allosteric regulation is given as [2]
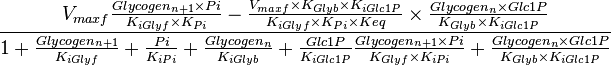
Parameter values
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
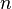
|
2 | Dimensionless | Recombinant, human muscle | |
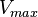
|
50 | 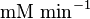
| ||
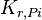
|
2.08 | mM | ||
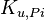
|
4.32 | mM | ||
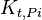
|
41.53 | mM | ||
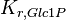
|
0.67 | mM | ||
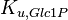
|
82.02 | mM | ||
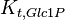
|
27.92 | mM | ||
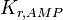
|
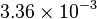
|
mM | ||
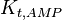
|
0.53 | mM | ||
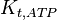
|
3.9 | mM | ||
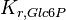
|
7.42 | mM | ||
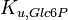
|
0.56 | mM | ||
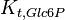
|
0.27 | mM | ||
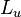
|
5.93 | Dimensionless | ||
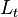
|
34741 | Dimensionless |
The parameter for the alternative equation are
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
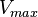
|
50 | 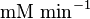
|
Rabbit | |
| Failed to parse (Cannot store math image on filesystem.): Ka | 4 | mM | ||
| Failed to parse (Cannot store math image on filesystem.): Kb | 1.7 | mM | ||
| Failed to parse (Cannot store math image on filesystem.): Kp | 2.7 | mM | ||
| Failed to parse (Cannot store math image on filesystem.): Kq | 0.15 | mM | ||
| Failed to parse (Cannot store math image on filesystem.): Ki_1 | 2 | mM | ||
| Failed to parse (Cannot store math image on filesystem.): Ki_2 | 4.7 | mM | ||
| Failed to parse (Cannot store math image on filesystem.): Ki_3 | 10.1 | mM | ||
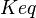
|
0.42 | mM |