Difference between revisions of "Metabolite concentrations"
| Line 1: | Line 1: | ||
| + | ==Initial Concentration== | ||
Initial concentration of the metabolites are listed below | Initial concentration of the metabolites are listed below | ||
| Line 94: | Line 95: | ||
|Glycogen | |Glycogen | ||
|112 <ref name="Lambeth_2002"></ref> | |112 <ref name="Lambeth_2002"></ref> | ||
| + | |- | ||
| + | |PHP | ||
| + | |0.60 <ref name = "Smallbone_2013">Smallbone K, Stanford NJ (2013). ''Kinetic modeling of metabolic pathways: Application to serine biosynthesis''. In: Systems Metabolic Engineering, Humana Press. pp. 113–121</ref> | ||
| + | |- | ||
| + | |PSER | ||
| + | |0.09 <ref name = "Smallbone_2013"></ref> | ||
| + | |- | ||
| + | |Serine | ||
| + | |4.90 <ref name = "Smallbone_2013"></ref> | ||
|} | |} | ||
Revision as of 16:27, 27 March 2014
Initial Concentration
Initial concentration of the metabolites are listed below
| Metabolites | Initial concentrations |
|---|---|
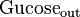
|
5 mM (Fixed) [1] |
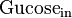
|
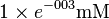 [2] [2]
|
| ATP | 8.70 mM [2] |
| Glc-6-P | 1.3 mM [2] |
| ADP | 2.7 mM [2] |
| Fru-6-P | 0.5 mM [2] |
| Fru-1,6-BP | 0.38 mM [2] |
| DHAP | 0.93 mM [2] |
| Gly3P | 0.9 mM [2] |
| NAD | 1.3 mM [2] |
| 1,3BPG | 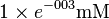 [2] [2]
|
| NADH | 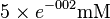 [2] [2]
|
| 3PG | 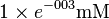 [2] [2]
|
| 2PG | 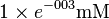 [2] [2]
|
| PEP | 0.32 mM [2] |
| Pyruvate | 8.5 mM [2] |
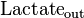
|
33 mM [2] (Fixed) |
| Glycogen | 135 mM [2] (Fixed) |
| Pi | 4 mM [2] (Fixed) |
| AMP | 0.4 mM [2] |
| 6PG | 0.39 mM [2] (Fixed) |
| Xy5P | 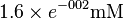 [3] (Fixed) [3] (Fixed)
|
| Ery4P | 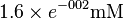 [3] (Fixed) [3] (Fixed)
|
| CIT | 1.7 [2] (Fixed) |
| F-2,6-BP | 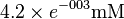 [2] (Fixed) [2] (Fixed)
|
| Glc-1-P | 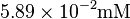 [4] [4]
|
| UTP | 0.13 [5] |
| PPi | 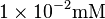 [6] [6]
|
| UDPG | 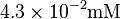 [7] [7]
|
| Glycogen | 112 [4] |
| PHP | 0.60 [8] |
| PSER | 0.09 [8] |
| Serine | 4.90 [8] |
References
- ↑ Marín-Hernández A , Rodríguez-Enríquez S, Vital-González P A, et al. (2006). Determining and understanding the control of glycolysis in fast-growth tumor cells. Flux control by an over-expressed but strongly product-inhibited hexokinase. FEBS J., 273 , pp. 1975–1988(doi)
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 2.18 2.19 2.20 2.21 Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011). Modeling cancer glycolysis. Biochim Biophys Acta, 1807:755–767 (doi)
- ↑ 3.0 3.1 Reitzer L J (1980). The pentose cycle. J. Biol. Chem., 255 , pp. 5616–5626
- ↑ 4.0 4.1 Lambeth MJ & Kushmerick MJ (2002). A computational model for glycogenolysis in skeletal muscle. Ann Biomed Eng 30, 808–827
- ↑ Keppler D, Rudiger J & Decker K (1970) Enzymatic determination of uracil nucleotides in tissues. Anal Biochem 38, 105–114.
- ↑ Palm, D.C. (2013). The regulatory design of glycogen metabolism in mammalian skeletal muscle (Ph.D.). University of Stellenbosch
- ↑ Albe KR, Butler MH & Wright BE (1990). Cellular concentrations of enzymes and their substrates. J Theor Biol 143, 163–195.
- ↑ 8.0 8.1 8.2 Smallbone K, Stanford NJ (2013). Kinetic modeling of metabolic pathways: Application to serine biosynthesis. In: Systems Metabolic Engineering, Humana Press. pp. 113–121