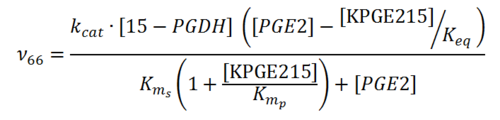Difference between revisions of "Transformation of PGE2 to 15-Keto-PGE2"
| Line 1: | Line 1: | ||
[[Welcome to the In-Silico Model of Cutaneous Lipids Wiki | Return to overview]] | [[Welcome to the In-Silico Model of Cutaneous Lipids Wiki | Return to overview]] | ||
| − | 15-hydroxyprostaglandin dehydrogenase, also known as (15-PGDH) metabolises PG into the 15-Keto variant via | + | 15-hydroxyprostaglandin dehydrogenase, also known as (15-PGDH) metabolises PG into the 15-Keto variant via the oxidation of 15(S)-hydroxyl group. The enzyme is constituently expressed in the skin (Finhelm1982) and within the eicosanoid network it is reported as metabolising PGE2 and PGF2a. |
It should be noted that Judson et al found that 15-PGDH expression of the enzyme decreases in response to UVR (Judson2010). | It should be noted that Judson et al found that 15-PGDH expression of the enzyme decreases in response to UVR (Judson2010). | ||
Revision as of 13:31, 1 February 2019
15-hydroxyprostaglandin dehydrogenase, also known as (15-PGDH) metabolises PG into the 15-Keto variant via the oxidation of 15(S)-hydroxyl group. The enzyme is constituently expressed in the skin (Finhelm1982) and within the eicosanoid network it is reported as metabolising PGE2 and PGF2a.
It should be noted that Judson et al found that 15-PGDH expression of the enzyme decreases in response to UVR (Judson2010).
Reaction
Catalysed by 15-PGDH
Chemical equation
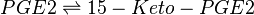
Rate equation
15-PGDH Parameters
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 0.008 | 
|
Rat Skin | Method:
pH:7.4 Temperature: 37'C Substrate: PGE2 + NAD+ |
[1] |
| 0.0075 | 
|
Rat Skin | Method:
pH:7.4 Temperature: 37'C Substrate: PGE2 + NADP+ |
[1] |
| 0.024 | 
|
Rat Skin | Method:
pH:7.4 Temperature: 37'C Substrate: PGF2a + NAD+ |
[1] |
| 0.023 | 
|
Rat Skin | Method:
pH:7.4 Temperature: 37'C Substrate: PGF2a + NADP+ |
[1] |
| 0.0039 | 
|
Purified Human 15-PGDH | Method: In vitro
Expression Vector: E. coli pH:7.5 Temperature: 37'C Substrate: PGE2 |
[2] |
| 0.0099 | 
|
Purified Rat 15-PGDH | Method: In vitro
Expression Vector: E. coli pH:7.5 Temperature: 37'C Substrate: PGE2 |
[2] |
| 0.0055 ± 0.0006 | 
|
Human | Method:In vitro
Expression Vector: E. Coli pH:8 Temperature: 25 Substrate: PGE2 + NAD+ |
[3] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 816 ± 18 | 
|
Human | Method: In vitro
Expression Vector: E. Coli pH:8 Temperature: 25 Substrate: PGE2 + NAD+ |
[3] |
| 366 ± 12 - 846 ± 12 | 
|
Human | Method: In vitro
Expression Vector: E. Coli pH:8 Temperature: 25 Substrate: PGE2 + NAD+ + Inhibitor |
[3] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 69.8 | 
|
Human | Expression Vector: Esophagus
Enzyme: 15-PGDH pH: 7.5 Temperature: 37 °C |
[4] |
| 22.1 | 
|
Human | Expression Vector: Heart
Enzyme: 15-PGDH pH: 7.5 Temperature: 37 °C |
[5] |
| 11.2 | 
|
Human | Expression Vector: Esophagus
Enzyme: 15-PGDH pH: 7.5 Temperature: 37 °C |
[5] |
| 7.68 | 
|
Human | Expression Vector: Skin
Enzyme: 15-PGDH pH: 7.5 Temperature: 37 °C |
Paxdb - Unknown |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| (-0.46818542) | kcal/mol | Calculated | Estimated
Enzyme: 15-PGDH Substrate: PGE2 Product: 15-dehydro-PGE2 pH: 7.3 ionic strength: 0.25 |
[6] |
References
- ↑ 1.0 1.1 1.2 1.3 N. Fincham, Novel prostaglandin dehydrogenase in rat skin. Biochem J. 1983 Apr 15;212(1):129-34.
- ↑ 2.0 2.1 Zhou H., C-Terminal region of human NAD+-dependent 15-hydroxyprostaglandin dehydrogenase is involved in the interaction with prostaglandin substrates. Eur J Biochem. 2001 Jun;268(12):3368-74.
- ↑ 3.0 3.1 3.2 F. Niesen,, High-Affinity Inhibitors of Human NAD+-Dependent 15-Hydroxyprostaglandin Dehydrogenase: Mechanisms of Inhibition and Structure-Activity Relationships PLoS One. 2010 Nov 2;5(11):e13719.
- ↑ M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587
- ↑ 5.0 5.1 M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471