Difference between revisions of "Limonene Synthase"
Aliah.hawari (talk | contribs) (→Kinetic Parameter Values) |
Aliah.hawari (talk | contribs) (→Parameterisation) |
||
| (23 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
You can go back to main page of the kinetic model [http://www.systemsbiology.ls.manchester.ac.uk/wiki/index.php/Kinetic_Model_of_Monoterpenoid_Biosynthesis_Wiki '''here''']. | You can go back to main page of the kinetic model [http://www.systemsbiology.ls.manchester.ac.uk/wiki/index.php/Kinetic_Model_of_Monoterpenoid_Biosynthesis_Wiki '''here''']. | ||
| − | + | Limonene synthase (LIMS) is not native to ''E. coli'' and a heterologous LIMS gene from ''Mentha spicata'' was engineered into the cell <ref name="AlonsoGutierrez2013">[https://www.sciencedirect.com/science/article/pii/S109671761300058X Alonso-Gutierrez, J., et al.](2013). "Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production." Metabolic Engineering 19: 33-41.</ref>. | |
| − | |||
:<math> | :<math> | ||
| Line 11: | Line 10: | ||
| + | == Reaction rate == | ||
| − | + | The reaction rate for LIMS is modelled using the reversible Michaelis-Menten equation <ref name="Sauro2011">Sauro, H. M. (2011). Appendix B: List of Common Rate Laws. Enzyme Kinetics for Systems Biology. United States of America, Future Skills Software: 279-290.</ref>, and is shown below: | |
| − | |||
| − | The reversible Michaelis-Menten equation | ||
:<math> | :<math> | ||
| − | V_\mathrm{LimSynth} = | + | V_\mathrm{LimSynth} = Kcat_\mathrm{LIMS}*[LIMS] * \cfrac {\cfrac{[GPP]}{Km_\mathrm{GPP}} * \left ( 1 - \cfrac {[Limonene]*[PP]}{[GPP]*K_\mathrm{eq}} \right )}{1 + \cfrac {[GPP]}{Km_\mathrm{GPP}} + \cfrac {[Limonene]}{Km_\mathrm{Limonene}} + \cfrac {[PP]}{Km_\mathrm{PP}} + \cfrac {[Limonene]*[PP]}{Km_\mathrm{Limonene}*Km_\mathrm{PP}}} |
</math> | </math> | ||
| Line 28: | Line 26: | ||
! scope="col" style="width: 50px; background: #ADD8E6;" | Units | ! scope="col" style="width: 50px; background: #ADD8E6;" | Units | ||
|- | |- | ||
| − | | V<sub>LimSynth</sub> || Reaction rate for Limonene Synthase || | + | | V<sub>LimSynth</sub> || Reaction rate for Limonene Synthase || μM/min |
|- | |- | ||
| − | | | + | | K<sub>cat<sub>LIMS</sub></sub> || Turnover number for limonene synthase || min<sup>-1</sup> |
|- | |- | ||
| − | | Km<sub>GPP</sub> || Michaelis-Menten constant for GPP || | + | | Km<sub>GPP</sub> || Michaelis-Menten constant for GPP || μM |
|- | |- | ||
| − | | Km<sub>Limonene</sub> || Michaelis-Menten constant for Limonene || | + | | Km<sub>Limonene</sub> || Michaelis-Menten constant for Limonene ||μM |
|- | |- | ||
| − | | Km<sub>PP</sub> || Michaelis-Menten constant for PP || | + | | Km<sub>PP</sub> || Michaelis-Menten constant for PP || μM |
|- | |- | ||
| − | | K<sub>eq</sub> || Equilibrium constant || | + | | K<sub>eq</sub> || Equilibrium constant || dimensionless |
|- | |- | ||
| − | | [GPP] || GPP concentration || | + | | [GPP] || GPP concentration || μM |
|- | |- | ||
| − | | [Limonene] || Limonene concentration || | + | | [Limonene] || Limonene concentration || μM |
|- | |- | ||
| − | | [PP]|| PP concentration || | + | | [PP]|| PP concentration || μM |
|} | |} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== Parameterisation == | == Parameterisation == | ||
| − | + | The parameterisation of LIMS for this model was performed according to our DIPPER protocol <ref name="Tsig2018>[https://www.nature.com/articles/s41596-018-0056-z Tsigkinopoulou, A., et al.] (2018). "Defining informative priors for ensemble modeling in systems biology." Nature protocols 13: 2643-2663.</ref>. Here, we list the thermodynamic and kinetic paramater values obtained from the literature, weights assigned and the log-normal distribution parameter values calculated. | |
| − | = | + | ===Thermodynamic parameter values=== |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Gibbs free energy values for LIMS are obtained from MetaCyc [http://biocyc.org/META/new-image?object=4.2.3.16-RXN (EC 4.2.3.16)] is -28.049988 kcal/mol <ref name="Latendresse2013"> Latendresse M. (2013). [http://www.biocyc.org/PGDBConceptsGuide.shtml#gibbs. "Computing Gibbs Free Energy of Compounds and Reactions in MetaCyc."] </ref> and Equilibrator [http://equilibrator.weizmann.ac.il/]. Table 2 summarizes the Δ<sub>r</sub>G° values found for LIMS and the calculated Keq. These Keq values are given an arbitrary equal weight of 1 as the Δ<sub>r</sub>G° values obtained were calculated from a group contribution method and do not have any measurement conditions that would allow us to assess the Keq values according to the weighting scheme set out in (insert link here). | Gibbs free energy values for LIMS are obtained from MetaCyc [http://biocyc.org/META/new-image?object=4.2.3.16-RXN (EC 4.2.3.16)] is -28.049988 kcal/mol <ref name="Latendresse2013"> Latendresse M. (2013). [http://www.biocyc.org/PGDBConceptsGuide.shtml#gibbs. "Computing Gibbs Free Energy of Compounds and Reactions in MetaCyc."] </ref> and Equilibrator [http://equilibrator.weizmann.ac.il/]. Table 2 summarizes the Δ<sub>r</sub>G° values found for LIMS and the calculated Keq. These Keq values are given an arbitrary equal weight of 1 as the Δ<sub>r</sub>G° values obtained were calculated from a group contribution method and do not have any measurement conditions that would allow us to assess the Keq values according to the weighting scheme set out in (insert link here). | ||
| Line 145: | Line 71: | ||
|- | |- | ||
|} | |} | ||
| − | |||
| − | |||
=== Kinetic Parameter Values === | === Kinetic Parameter Values === | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{| class="wikitable" | {| class="wikitable" | ||
| − | |+ Table 2: | + | |+ Table 2: Km for GPP values for LIMS |
| − | + | | align="center" style="background:#f0f0f0;"|'''Parameter''' | |
| − | + | | align="center" style="background:#f0f0f0;"|'''Value''' | |
| − | + | | align="center" style="background:#f0f0f0;"|'''Error''' | |
| − | + | | align="center" style="background:#f0f0f0;"|'''Weight''' | |
| + | | align="center" style="background:#f0f0f0;"|'''Error type''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Description''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''References''' | ||
| + | |- | ||
| + | | Km_gpp_LIMS||47.4||3.8||128||0||LIMS gene from Lavandula angustifolia was expressed in E. coli. The kinetics were measured in vitro at 30°C||<ref name="Landmann2007">[https://www.sciencedirect.com/science/article/pii/S0003986107003013 Landmann, C., et al.] (2007). "Cloning and characterization of three terpene synthases from lavender (''Lavandula angustifolia'')." Archives of Biochemistry and Biophysics 465: 417-429.</ref> | ||
|- | |- | ||
| − | | | + | | Km_gpp_LIMS||130||NaN||32||0||LIMS gene from Citrus sinensis (orange) was expressed in E. coli. The Km was measured in vitro at 20°C.||<ref name="Entova2013">[Entova, S.](2013). Kinetic characterization, crystallization, and photosynthetic expression of (+)-$R-limonene synthase from C. sinensis. Department of Biochemistry. Massachusetts, Brandeis University. Master's: 55.</ref> |
| − | | | ||
| − | |||
| − | | | ||
|- | |- | ||
| − | | | + | | Km_gpp_LIMS||6.8||NaN||32||0||LIMS gene from Cannabis sativa L. var. 'Skunk' plants was expressed in E. coli. The kinetics were measured at 40°C.||<ref name="Gunnewich2007"> [https://www.researchgate.net/publication/42089356_Functional_expression_and_characterization_of_trichome-specific_--limonene_synthase_and_-a-pinene_synthase_from_Cannabis_sativa Günnewich, N., Page, J.E., Köllner, T.G., Degenhardt, J., & Kutchan, T.M] 2007. "Functional expression and characterization of trichome-specific (-)-limonene synthase and (+)-α-pinene synthase from ''Cannabis sativa'' ". Nat. Prod. Comm. 2(3): 223-232. </ref> |
| − | | | ||
| − | | | ||
|- | |- | ||
| − | | | + | | Km_gpp_LIMS||0.7||NaN||128||0||LIMS gene from Citrus limon (lemon) was expressed in E. col. Kinetics were measured in vitro at 30°C.||<ref name="Lucker2002">[https://febs.onlinelibrary.wiley.com/doi/full/10.1046/j.1432-1033.2002.02985.x Lücker, J., et al.] (2002). "Monoterpene biosynthesis in lemon (''Citrus limon'') cDNA isolation and functional analysis of four monoterpene synthases." Eur. J. Biochem. 269: 3160-3171. |
| − | | | + | </ref> |
| − | |||
| − | | | ||
|- | |- | ||
| − | | | + | | Km_gpp_LIMS||1.25||NaN||16||0||LIMS isolated from Ricciocarpos natans. Kinetics measured at 32°C and pH7.0. ||<ref name="Adam1996">[https://www.sciencedirect.com/science/article/pii/S0003986196903525 Adam, K.-P., et al.] (1996). "Partial purification and characterization of a monoterpene cyclase, limonene synthase, from the liverwort ''Ricciocarpus natans''." Archives of Biochemistry and Biophysics 332(2): 352-356. </ref> |
| − | | | ||
| − | | | ||
|- | |- | ||
| − | | | + | | Km_gpp_LIMS||1.8||NaN||32||0||LIMS isolated from Mentha x piperita (peppermint). Kinetics were measured at 30°C.||<ref name="Rajaonarivony1992">[https://www.sciencedirect.com/science/article/pii/0003986192905436 Rajaonarivony, J. I. M., et al.] (1992). "Characterization and mechanism of (4S)-limonene synthase, a monoterpene cyclase from the glandular trichomes of peppermint (Mentha X piperita)." Archives of Biochemistry and Biophysics 296(1): 49-57.</ref> |
| − | | style=" | + | |} |
| − | | style=" | + | |
| − | | | + | {| class="wikitable" |
| + | |+ Table 3: Turnover number values for LIMS | ||
| + | | align="center" style="background:#f0f0f0;"|'''Parameter''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Value''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Error''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Weight''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Error type''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Description''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''References''' | ||
|- | |- | ||
| − | | | + | | Kcat_LIMS||0.72||NaN||128||0||LIMS gene from Lavandula angustifolia was expressed in E. coli. The kinetics were measured in vitro at 30°C||<ref name="Landmann2007" /> |
| − | | 0. | ||
| − | | | ||
| − | | | ||
|- | |- | ||
| − | | | + | | Kcat_LIMS||7.8||NaN||32||0||LIMS gene from Citrus sinensis (orange) was expressed in E. coli. The enzyme activities were measured in vitro at 20°C and 37°C.||<ref name="Entova2013" /> |
| − | | 0 | ||
| − | | | ||
| − | | | ||
| − | | | ||
|- | |- | ||
| − | | | + | | Kcat_LIMS||2.4||NaN||32||0||LIMS gene from Citrus sinensis (orange) was expressed in E. coli. The enzyme activities were measured in vitro at 20°C and 37°C.||<ref name="Entova2013" /> |
| − | | | ||
| − | | | ||
| − | | | ||
| − | |||
| − | |||
| − | | | ||
|- | |- | ||
| − | | | + | | Kcat_LIMS||4.92||NaN||32||0||LIMS gene from Cannabis sativa L. var. 'Skunk' plants was expressed in E. coli. The kinetics were measured at 40°C.||<ref name="Gunnewich2007" /> |
| − | | 0 | ||
| − | | | ||
| − | | | ||
| − | |||
| − | | | ||
|- | |- | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| − | === | + | === BRENDA data === |
| + | To further enrich the kinetic parameter values for LIMS, parameter values from EC 4 to EC 4.2.3.* that can be obtained from BRENDA is downloaded. These BRENDA data is integrated with the rest of the kinetic parameter values using our ‘BRENDA Add-on’ protocol. In the BRENDA Add-On protocol, we’ve specified six different ‘EC cases’ that are arranged in order of rank. These EC cases are essentially six different datasets of parameter values downloaded from BRENDA that are filtered according to the specific enzyme class and organism of interest. For this case study example, six different ‘EC case’ datasets were downloaded from BRENDA each for Km and Kcat parameters (Tables below). | ||
| − | {| class="wikitable | + | {| class="wikitable" |
| + | |+ Table 7: Input matrix for Km values from BRENDA for EC 4.2.3.16 | ||
| + | |+ align="bottom" style="caption-side: bottom; text-align: left;" |Parameter value and uncertainty is in µM. Each row represents EC case 2 – 6 respectively. EC case 1 is discarded as number of values obtained were <50. . | ||
| + | |+ align="bottom" style="caption-side: bottom; text-align: left;" |*Uncertainty values in this table correspond to the standard deviation calculated from SEM reported in literature | ||
| + | |+ align="bottom" style="caption-side: bottom; text-align: left;" |** Uncertainty type is multiplicative, therefore ‘1’ is defined. | ||
| + | |||
| + | ! style="font-weight: bold;" | Parameter value | ||
| + | ! style="font-weight: bold;" | Uncertainty* | ||
| + | ! style="font-weight: bold;" | Weight | ||
| + | ! style="font-weight: bold;" | Uncertainty type** | ||
|- | |- | ||
| − | + | | 6.80 | |
| − | + | | 14.9566 | |
| − | + | | 11.9024 | |
| − | + | | 1 | |
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | 49 |
| − | | | + | | 69.0555 |
| − | + | | 7.4390 | |
| − | | | + | | 1 |
| − | | | ||
| − | |||
|- | |- | ||
| − | | | + | | 121 |
| − | | | + | | 36.1126 |
| − | + | | 2.9756 | |
| − | + | | 1 | |
| − | | | ||
| − | | | ||
|- | |- | ||
| − | | | + | | 500 |
| − | + | | 36.9586 | |
| − | + | | 1.4878 | |
| − | + | | 1 | |
| − | |||
| − | |||
| − | | | ||
| − | |||
| − | | | ||
| − | |||
| − | |||
| − | |||
| − | | | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|- | |- | ||
| + | | 300 | ||
| + | | 23.8770 | ||
| + | | 0.5951 | ||
| + | | 1 | ||
|} | |} | ||
| − | + | {| class="wikitable" | |
| − | {| class="wikitable | + | |+ Input matrix for K<sub>cat</sub> values from BRENDA for EC 4.2.3.16 |
| − | + | |+ align="bottom" style="caption-side: bottom; text-align: left;" |Parameter value and uncertainty is in µM. Each row represents EC case 2 – 6 respectively. EC case 1 is discarded as number of values obtained were <50. . | |
| + | |+ align="bottom" style="caption-side: bottom; text-align: left;" |*Uncertainty values in this table correspond to the standard deviation calculated from SEM reported in literature | ||
| + | |+ align="bottom" style="caption-side: bottom; text-align: left;" |** Uncertainty type is multiplicative, therefore ‘1’ is defined. | ||
| + | ! style="font-weight: bold;" | Parameter value | ||
| + | ! style="font-weight: bold;" | Uncertainty* | ||
| + | ! style="font-weight: bold;" | Weight | ||
| + | ! style="font-weight: bold;" | Uncertainty type** | ||
|- | |- | ||
| − | | | + | | 2.040 |
| + | | 38.2439 | ||
| + | | 9.5610 | ||
| + | | 1 | ||
|- | |- | ||
| − | | | + | | 17.400 |
| + | | 109.1960 | ||
| + | | 5.9756 | ||
| + | | 1 | ||
|- | |- | ||
| − | | | + | | 200.698 |
| + | | 177.0428 | ||
| + | | 2.3902 | ||
| + | | 1 | ||
|- | |- | ||
| − | | | + | | 192.000 |
| + | | 44.6715 | ||
| + | | 1.1951 | ||
| + | | 1 | ||
|- | |- | ||
| − | | | + | | 168.000 |
| − | | | + | | 62.4126 |
| − | + | | 0.4780 | |
| − | + | | 1 | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| − | === | + | === Log-normal distribution parameters === |
| − | The | + | The Mode, Confidence Interval (CI) factor, mu and sigma for each parameter distribution is calculated using our scripts as detailed in the DIPPER protocol <ref name="Tsig2018>[https://www.nature.com/articles/s41596-018-0056-z Tsigkinopoulou, A., et al.] (2018). "Defining informative priors for ensemble modeling in systems biology." Nature protocols 13: 2643-2663.</ref>. |
| − | {| class="wikitable" style=" | + | {| class="wikitable" |
| + | | align="center" style="background:#f0f0f0;"|'''Parameter''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Brenda Add-on''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Mode''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''CI Factor''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Mu''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Sigma''' | ||
| + | |- | ||
| + | | Km_gpp_LIMS||✘||6.8||7.6504||3.3719||1.2062 | ||
|- | |- | ||
| − | + | | Kcat_LIMS||✘||0.8001||2.6307||0.3424||0.7519 | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | | | + | | Km_gpp_LIMS||✔|||||||| |
|- | |- | ||
| − | | | + | | Kcat_LIMS||✔|||||||| |
|- | |- | ||
| − | |||
|} | |} | ||
| − | === | + | === Log-normal distribution parameter estimation === |
This section can be found [[ LimSynth: Parameter Estimation | ''' HERE ''']] | This section can be found [[ LimSynth: Parameter Estimation | ''' HERE ''']] | ||
Latest revision as of 14:14, 29 November 2018
You can go back to main page of the kinetic model here.
Limonene synthase (LIMS) is not native to E. coli and a heterologous LIMS gene from Mentha spicata was engineered into the cell [1].
Contents
Reaction rate
The reaction rate for LIMS is modelled using the reversible Michaelis-Menten equation [2], and is shown below:
where :
| Parameter | Description | Units |
|---|---|---|
| VLimSynth | Reaction rate for Limonene Synthase | μM/min |
| KcatLIMS | Turnover number for limonene synthase | min-1 |
| KmGPP | Michaelis-Menten constant for GPP | μM |
| KmLimonene | Michaelis-Menten constant for Limonene | μM |
| KmPP | Michaelis-Menten constant for PP | μM |
| Keq | Equilibrium constant | dimensionless |
| [GPP] | GPP concentration | μM |
| [Limonene] | Limonene concentration | μM |
| [PP] | PP concentration | μM |
Parameterisation
The parameterisation of LIMS for this model was performed according to our DIPPER protocol [3]. Here, we list the thermodynamic and kinetic paramater values obtained from the literature, weights assigned and the log-normal distribution parameter values calculated.
Thermodynamic parameter values
Gibbs free energy values for LIMS are obtained from MetaCyc (EC 4.2.3.16) is -28.049988 kcal/mol [4] and Equilibrator [1]. Table 2 summarizes the ΔrG° values found for LIMS and the calculated Keq. These Keq values are given an arbitrary equal weight of 1 as the ΔrG° values obtained were calculated from a group contribution method and do not have any measurement conditions that would allow us to assess the Keq values according to the weighting scheme set out in (insert link here).
| ΔrG°(kcal/mol) | Keq | Error (±) | Source | Weight |
|---|---|---|---|---|
| -28.050 | 3.843E+20 | N/A | MetaCyc | 1 |
| -42.161 ± 2.844 | 8.711E+30 | 5.876E+29 | Equilibrator | 1 |
Kinetic Parameter Values
| Parameter | Value | Error | Weight | Error type | Description | References |
| Km_gpp_LIMS | 47.4 | 3.8 | 128 | 0 | LIMS gene from Lavandula angustifolia was expressed in E. coli. The kinetics were measured in vitro at 30°C | [5] |
| Km_gpp_LIMS | 130 | NaN | 32 | 0 | LIMS gene from Citrus sinensis (orange) was expressed in E. coli. The Km was measured in vitro at 20°C. | [6] |
| Km_gpp_LIMS | 6.8 | NaN | 32 | 0 | LIMS gene from Cannabis sativa L. var. 'Skunk' plants was expressed in E. coli. The kinetics were measured at 40°C. | [7] |
| Km_gpp_LIMS | 0.7 | NaN | 128 | 0 | LIMS gene from Citrus limon (lemon) was expressed in E. col. Kinetics were measured in vitro at 30°C. | [8] |
| Km_gpp_LIMS | 1.25 | NaN | 16 | 0 | LIMS isolated from Ricciocarpos natans. Kinetics measured at 32°C and pH7.0. | [9] |
| Km_gpp_LIMS | 1.8 | NaN | 32 | 0 | LIMS isolated from Mentha x piperita (peppermint). Kinetics were measured at 30°C. | [10] |
| Parameter | Value | Error | Weight | Error type | Description | References |
| Kcat_LIMS | 0.72 | NaN | 128 | 0 | LIMS gene from Lavandula angustifolia was expressed in E. coli. The kinetics were measured in vitro at 30°C | [5] |
| Kcat_LIMS | 7.8 | NaN | 32 | 0 | LIMS gene from Citrus sinensis (orange) was expressed in E. coli. The enzyme activities were measured in vitro at 20°C and 37°C. | [6] |
| Kcat_LIMS | 2.4 | NaN | 32 | 0 | LIMS gene from Citrus sinensis (orange) was expressed in E. coli. The enzyme activities were measured in vitro at 20°C and 37°C. | [6] |
| Kcat_LIMS | 4.92 | NaN | 32 | 0 | LIMS gene from Cannabis sativa L. var. 'Skunk' plants was expressed in E. coli. The kinetics were measured at 40°C. | [7] |
BRENDA data
To further enrich the kinetic parameter values for LIMS, parameter values from EC 4 to EC 4.2.3.* that can be obtained from BRENDA is downloaded. These BRENDA data is integrated with the rest of the kinetic parameter values using our ‘BRENDA Add-on’ protocol. In the BRENDA Add-On protocol, we’ve specified six different ‘EC cases’ that are arranged in order of rank. These EC cases are essentially six different datasets of parameter values downloaded from BRENDA that are filtered according to the specific enzyme class and organism of interest. For this case study example, six different ‘EC case’ datasets were downloaded from BRENDA each for Km and Kcat parameters (Tables below).
| Parameter value | Uncertainty* | Weight | Uncertainty type** |
|---|---|---|---|
| 6.80 | 14.9566 | 11.9024 | 1 |
| 49 | 69.0555 | 7.4390 | 1 |
| 121 | 36.1126 | 2.9756 | 1 |
| 500 | 36.9586 | 1.4878 | 1 |
| 300 | 23.8770 | 0.5951 | 1 |
| Parameter value | Uncertainty* | Weight | Uncertainty type** |
|---|---|---|---|
| 2.040 | 38.2439 | 9.5610 | 1 |
| 17.400 | 109.1960 | 5.9756 | 1 |
| 200.698 | 177.0428 | 2.3902 | 1 |
| 192.000 | 44.6715 | 1.1951 | 1 |
| 168.000 | 62.4126 | 0.4780 | 1 |
Log-normal distribution parameters
The Mode, Confidence Interval (CI) factor, mu and sigma for each parameter distribution is calculated using our scripts as detailed in the DIPPER protocol [3].
| Parameter | Brenda Add-on | Mode | CI Factor | Mu | Sigma |
| Km_gpp_LIMS | ✘ | 6.8 | 7.6504 | 3.3719 | 1.2062 |
| Kcat_LIMS | ✘ | 0.8001 | 2.6307 | 0.3424 | 0.7519 |
| Km_gpp_LIMS | ✔ | ||||
| Kcat_LIMS | ✔ |
Log-normal distribution parameter estimation
This section can be found HERE
Simulations
Simulations performed can be found HERE.
References
- ↑ Alonso-Gutierrez, J., et al.(2013). "Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production." Metabolic Engineering 19: 33-41.
- ↑ Sauro, H. M. (2011). Appendix B: List of Common Rate Laws. Enzyme Kinetics for Systems Biology. United States of America, Future Skills Software: 279-290.
- ↑ 3.0 3.1 Tsigkinopoulou, A., et al. (2018). "Defining informative priors for ensemble modeling in systems biology." Nature protocols 13: 2643-2663.
- ↑ Latendresse M. (2013). "Computing Gibbs Free Energy of Compounds and Reactions in MetaCyc."
- ↑ 5.0 5.1 Landmann, C., et al. (2007). "Cloning and characterization of three terpene synthases from lavender (Lavandula angustifolia)." Archives of Biochemistry and Biophysics 465: 417-429.
- ↑ 6.0 6.1 6.2 [Entova, S.](2013). Kinetic characterization, crystallization, and photosynthetic expression of (+)-$R-limonene synthase from C. sinensis. Department of Biochemistry. Massachusetts, Brandeis University. Master's: 55.
- ↑ 7.0 7.1 Günnewich, N., Page, J.E., Köllner, T.G., Degenhardt, J., & Kutchan, T.M 2007. "Functional expression and characterization of trichome-specific (-)-limonene synthase and (+)-α-pinene synthase from Cannabis sativa ". Nat. Prod. Comm. 2(3): 223-232.
- ↑ Lücker, J., et al. (2002). "Monoterpene biosynthesis in lemon (Citrus limon) cDNA isolation and functional analysis of four monoterpene synthases." Eur. J. Biochem. 269: 3160-3171.
- ↑ Adam, K.-P., et al. (1996). "Partial purification and characterization of a monoterpene cyclase, limonene synthase, from the liverwort Ricciocarpus natans." Archives of Biochemistry and Biophysics 332(2): 352-356.
- ↑ Rajaonarivony, J. I. M., et al. (1992). "Characterization and mechanism of (4S)-limonene synthase, a monoterpene cyclase from the glandular trichomes of peppermint (Mentha X piperita)." Archives of Biochemistry and Biophysics 296(1): 49-57.
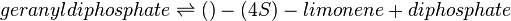
![V_\mathrm{LimSynth} = Kcat_\mathrm{LIMS}*[LIMS] * \cfrac {\cfrac{[GPP]}{Km_\mathrm{GPP}} * \left ( 1 - \cfrac {[Limonene]*[PP]}{[GPP]*K_\mathrm{eq}} \right )}{1 + \cfrac {[GPP]}{Km_\mathrm{GPP}} + \cfrac {[Limonene]}{Km_\mathrm{Limonene}} + \cfrac {[PP]}{Km_\mathrm{PP}} + \cfrac {[Limonene]*[PP]}{Km_\mathrm{Limonene}*Km_\mathrm{PP}}}](/wiki/images/math/b/c/3/bc34cd8a82726466dee4043dc9cc6bf8.png)