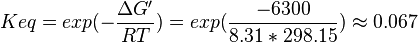Difference between revisions of "Glyceraldehyde-3-phosphate dehydrogenase"
(→Parameters with uncertainty) |
|||
| (6 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
Ordererd Ter-Bi reversible Michaelis-Menten equation for non-interacting substrates. <ref name="Hernandez2011"> Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 ([http://dx.doi.org/10.1016/j.bbabio.2010.11.006 doi]) </ref> | Ordererd Ter-Bi reversible Michaelis-Menten equation for non-interacting substrates. <ref name="Hernandez2011"> Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 ([http://dx.doi.org/10.1016/j.bbabio.2010.11.006 doi]) </ref> | ||
| − | <center><math>v = \frac{ V_{mf}\frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} - V_{mr} \frac{[1,3BPG][NADH]}{K_{1,3BPG}K_{NADH}} }{1 + \frac{[NAD]}{K_{NAD}} + \frac{[NAD][Gly3P]}{K_{NAD}K_{Gly3P}} + \frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} + \frac{[1,3BPG][NADH]}{K_{1,3BPG}K_{NADH}} | + | <center><math>v = \frac{ V_{mf}\frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} - V_{mr} \frac{[1,3BPG][NADH]}{K_{1,3BPG}K_{NADH}} }{1 + \frac{[NAD]}{K_{NAD}} + \frac{[NAD][Gly3P]}{K_{NAD}K_{Gly3P}} + \frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} + \frac{[1,3BPG][NADH]}{K_{1,3BPG}K_{NADH}} +\frac{[NADH]}{K_{NADH}} }</math></center> |
Modified rate law for taking Thermodynamic constant into consideration | Modified rate law for taking Thermodynamic constant into consideration | ||
| − | <center><math>v = \frac{ V_{mf}\frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} \left( 1 - \frac{[1,3BPG][NADH]}{K_{eq}[NAD][Gly3P][Pi]} }{1 + \frac{[NAD]}{K_{NAD}} + \frac{[NAD][Gly3P]}{K_{NAD}K_{Gly3P}} + \frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} + \frac{[1,3BPG][NADH]}{K_{1,3BPG}K_{NADH}} | + | <center><math>v = \frac{ V_{mf}\frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} \left( 1 - \frac{[1,3BPG][NADH]}{K_{eq}[NAD][Gly3P][Pi]} \right)}{1 + \frac{[NAD]}{K_{NAD}} + \frac{[NAD][Gly3P]}{K_{NAD}K_{Gly3P}} + \frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} + \frac{[1,3BPG][NADH]}{K_{1,3BPG}K_{NADH}} +\frac{[NADH]}{K_{NADH}} }</math></center> |
==Parameters== | ==Parameters== | ||
| Line 71: | Line 71: | ||
|- | |- | ||
|<math>V_{mf}</math> | |<math>V_{mf}</math> | ||
| − | |<math> 2 \pm 0.74 (5) </math> <ref name="Hernandez_2006"></ref> | + | |<math> 2 \pm 0.74 (5) </math> <ref name="Hernandez_2006"></ref> <br> <math> 130 \pm 48.1 (5) </math> <ref name="Hernandez_2006"></ref> |
| − | |<math> mM \times min^{-1} </math> | + | |<math>U\cdot(\text{mg protein})^{-1}</math> <br> <math> mM \times min^{-1} </math> |
|HeLa cell line | |HeLa cell line | ||
|rowspan="7"| | |rowspan="7"| | ||
| Line 106: | Line 106: | ||
|Normal tissue in human | |Normal tissue in human | ||
|} | |} | ||
| + | |||
| + | === Equilibrium constant === | ||
| + | {|class="wikitable" | ||
| + | ! Equilibrium constant | ||
| + | ! Conditions | ||
| + | ! Source | ||
| + | |- | ||
| + | | 0.078 | ||
| + | | pH=7, T=25°C | ||
| + | | Lehninger, (2008)<ref name="lehninger2008">David L. Nelson, Michael M. Cox (2008), Lehninger Principles of Biochemistry (5th edn), W. H. Freeman and Company</ref> p 553:<br/> | ||
| + | <math>\Delta G' = 6.3\ kJ.mol^{-1}</math>, <math>Keq = exp(-\frac{\Delta G'}{RT}) = exp(\frac{-6300}{8.31*298.15}) \approx 0.078</math> | ||
| + | |- | ||
| + | | 0.067 | ||
| + | | pH=7, T=25°C | ||
| + | | Voet et al.<ref name="voet">Voet, D., Voet., J.G. and Pratt, C. W. (1999) Fundamentals of biochemistry, Wiley</ref> from Newshole et al. (1973) <ref name="newshole73">Newshole, E.A. and Stuart, C. (1973) Regulation in Metabolism, Wiley</ref>p 97:<br/> | ||
| + | <math>\Delta G' = 6.7\ kJ.mol^{-1}</math>, <math>Keq = exp(-\frac{\Delta G'}{RT}) = exp(\frac{-6300}{8.31*298.15}) \approx 0.067</math> | ||
| + | |- | ||
| + | | 0.08 | ||
| + | | pH=7, T=298.15 Kelvin | ||
| + | | Corrected for pH from Cori et al. 1950 (NIST database<ref name="nist">Goldberg R.N., Tewari Y.B. and Bhat T.N. (2004) Bioinformatics 20(16):2874-2877 [[http://www.ncbi.nlm.nih.gov/pubmed?term=15145806 pmid: 15145806]]</ref> [[http://xpdb.nist.gov/enzyme_thermodynamics/enzyme_data1.pl?T1=50COR/VEL_252 50COR/VEL_252]]) pH=7.09 | ||
| + | |} | ||
| + | |||
| + | *The mean and Std. Dev. for Keq would be <math>0.075 \pm 0.007</math> | ||
==References== | ==References== | ||
<references/> | <references/> | ||
Latest revision as of 11:00, 16 October 2014
The Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) enzyme serves two functions in glycolytic pathway. First the enzyme transfers a hydrogen (H-) from glyceraldehyde phosphate (Gly3P) to the oxidizing agent Nicotinamide Adenine Dinucleotide (NAD+) to form NADH. Next it adds a phosphate (P) from the cytosol to the oxidized Gly3P to form 1, 3-bisphosphoglycerate.
Contents
Chemical equation
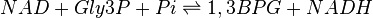
Rate equation
Ordererd Ter-Bi reversible Michaelis-Menten equation for non-interacting substrates. [1]
![v = \frac{ V_{mf}\frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} - V_{mr} \frac{[1,3BPG][NADH]}{K_{1,3BPG}K_{NADH}} }{1 + \frac{[NAD]}{K_{NAD}} + \frac{[NAD][Gly3P]}{K_{NAD}K_{Gly3P}} + \frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} + \frac{[1,3BPG][NADH]}{K_{1,3BPG}K_{NADH}} +\frac{[NADH]}{K_{NADH}} }](/wiki/images/math/8/1/3/813cf19acd739ad1b01dab007cce42bd.png)
Modified rate law for taking Thermodynamic constant into consideration
![v = \frac{ V_{mf}\frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} \left( 1 - \frac{[1,3BPG][NADH]}{K_{eq}[NAD][Gly3P][Pi]} \right)}{1 + \frac{[NAD]}{K_{NAD}} + \frac{[NAD][Gly3P]}{K_{NAD}K_{Gly3P}} + \frac{[NAD][Gly3P][Pi]}{K_{NAD}K_{Gly3P}K_{Pi}} + \frac{[1,3BPG][NADH]}{K_{1,3BPG}K_{NADH}} +\frac{[NADH]}{K_{NADH}} }](/wiki/images/math/e/5/3/e53e8bbfe1a674c134f439b46f5e59a5.png)
Parameters
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
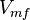
|
0.58 [2] | 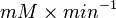
|
HeLa cell line | |
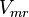
|
0.72[2] | 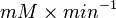
| ||
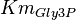
|
0.19[1] | mM | ||
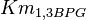
|
0.022[1] | mM | ||
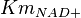
|
0.09[1] | mM | ||
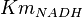
|
0.01[1] | mM | ||
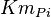
|
29[1] | mM |
Parameters with uncertainty
- Four values of
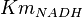 have been collected from Lambeir et. al. (1991)[3] for different organisms.
have been collected from Lambeir et. al. (1991)[3] for different organisms. 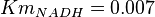 for Trypanosoma brucei,
for Trypanosoma brucei, 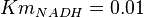 for Homo sapiens,
for Homo sapiens, 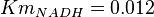 for Geobacillus stearothermophilus and
for Geobacillus stearothermophilus and 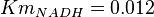 for Oryctolagus cuniculus. Calculating mean and std. dev. from these 4 values gives
for Oryctolagus cuniculus. Calculating mean and std. dev. from these 4 values gives 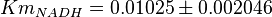
- Three values collected under different conditions from human cell is reported for
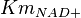 in Ryzlak (1998) et. al. [4]. Those values are 0.01, 0.02, 0.027. The mean and Std. Dev. from these three values are
in Ryzlak (1998) et. al. [4]. Those values are 0.01, 0.02, 0.027. The mean and Std. Dev. from these three values are 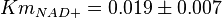
- Three values for
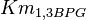 have also been reported under different conditions from human cell in Ryzlak (1998) et. al. [4]. Those values are 0.01, 0.012, 0.018. The mean and Std. Dev. from these three values are
have also been reported under different conditions from human cell in Ryzlak (1998) et. al. [4]. Those values are 0.01, 0.012, 0.018. The mean and Std. Dev. from these three values are 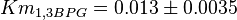
- Three values for
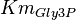 have also been reported under different conditions from human cell in Ryzlak (1998) et. al. [4]. Those values are 0.032, 0.035, 0.042. The mean and Std. Dev. from these three values are
have also been reported under different conditions from human cell in Ryzlak (1998) et. al. [4]. Those values are 0.032, 0.035, 0.042. The mean and Std. Dev. from these three values are 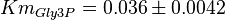
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
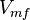
|
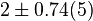 [2] [2] 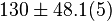 [2] [2]
|
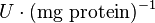 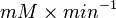
|
HeLa cell line | |
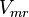
|
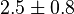 (5)[2] (5)[2]
|
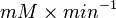
|
||
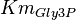
|
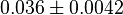
|
mM | Human brain | |
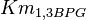
|
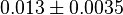
|
mM | Human brain | |
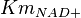
|
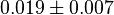
|
mM | Human brain | |
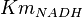
|
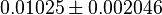
|
mM | Multiple organism | |
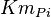
|
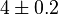 [5] [5]
|
mM | Normal tissue in human |
Equilibrium constant
| Equilibrium constant | Conditions | Source |
|---|---|---|
| 0.078 | pH=7, T=25°C | Lehninger, (2008)[6] p 553:
|
| 0.067 | pH=7, T=25°C | Voet et al.[7] from Newshole et al. (1973) [8]p 97:
|
| 0.08 | pH=7, T=298.15 Kelvin | Corrected for pH from Cori et al. 1950 (NIST database[9] [50COR/VEL_252]) pH=7.09 |
- The mean and Std. Dev. for Keq would be
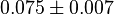
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 (doi)
- ↑ 2.0 2.1 2.2 2.3 2.4 Marín-Hernández A , Rodríguez-Enríquez S, Vital-González P A, et al. (2006). Determining and understanding the control of glycolysis in fast-growth tumor cells. Flux control by an over-expressed but strongly product-inhibited hexokinase. FEBS J., 273 , pp. 1975–1988(doi)
- ↑ Lambeir, A.M.; Loiseau, A.M.; Kuntz, D.A.; Vellieux, F.M.; Michels, P.A.M.; Opperdoes, F.R. (1991), The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic properties and comparison with homologous enzymes, Eur. J. Biochem. 198, 429-435
- ↑ 4.0 4.1 4.2 Ryzlak, M.T.; Pietruszko, R. (1998), Heterogeneity of glyceraldehyde-3-phosphate dehydrogenase from human brain, Biochim. Biophys. Acta 954, 309-324
- ↑ Patra, S.; Ghosh, S.; Bera, S.; Roy, A.; Ray, S.; Ray, M. (2009), Molecular characterization of tumor associated glyceraldehyde-3-phosphate dehydrogenase, Biochemistry (Moscow) 74, 717-727
- ↑ David L. Nelson, Michael M. Cox (2008), Lehninger Principles of Biochemistry (5th edn), W. H. Freeman and Company
- ↑ Voet, D., Voet., J.G. and Pratt, C. W. (1999) Fundamentals of biochemistry, Wiley
- ↑ Newshole, E.A. and Stuart, C. (1973) Regulation in Metabolism, Wiley
- ↑ Goldberg R.N., Tewari Y.B. and Bhat T.N. (2004) Bioinformatics 20(16):2874-2877 [pmid: 15145806]
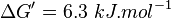 ,
, 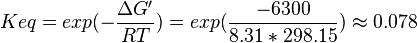
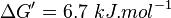 ,
,