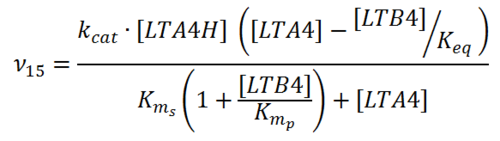Difference between revisions of "Transformation of LTA4 to LTB4"
| (4 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Welcome to the In-Silico Model of Cutaneous Lipids Wiki | Return to overview]] | [[Welcome to the In-Silico Model of Cutaneous Lipids Wiki | Return to overview]] | ||
| + | |||
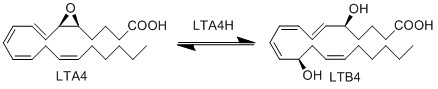
| + | LTA4 is subsequently enzymatically hydrolysed to LTB4 by LTA4 hydrolase (LTA4H). The enzyme performs this reaction by opening the epoxide and creating a carbocation intermediate. The charge of the carbocation delocalises over the triene system and is subsequently subject to nucleophilic attack by water at C12, resulting in the stereospecific addition of a hydroxyl group and the generation of LTB4. | ||
| + | |||
| + | |||
== Reaction == | == Reaction == | ||
| Line 14: | Line 18: | ||
== Parameters == | == Parameters == | ||
| + | === K<sub>ms</sub> === | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Literature values |
|- | |- | ||
! Value | ! Value | ||
| Line 21: | Line 26: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 30: | Line 36: | ||
pH:8 | pH:8 | ||
Temperature: 20 (Room Temperature) | Temperature: 20 (Room Temperature) | ||
| + | |128 | ||
|<ref name="Mueller1996"> [www.ncbi.nlm.nih.gov/pubmed/8650196 Mueller M. , "Leukotriene A4 hydrolase: protection from mechanism-based inactivation by mutation of tyrosine-378.'' Proc Natl Acad Sci U S A. 1996 Jun 11;93(12):5931-5.]</ref> | |<ref name="Mueller1996"> [www.ncbi.nlm.nih.gov/pubmed/8650196 Mueller M. , "Leukotriene A4 hydrolase: protection from mechanism-based inactivation by mutation of tyrosine-378.'' Proc Natl Acad Sci U S A. 1996 Jun 11;93(12):5931-5.]</ref> | ||
|- | |- | ||
| Line 39: | Line 46: | ||
pH:8 | pH:8 | ||
Temperature: 20 (Room Temperature) | Temperature: 20 (Room Temperature) | ||
| + | |16 | ||
|<ref name="Stromberg1998"> [www.ncbi.nlm.nih.gov/pubmed/9744798 Stromberg F. , "Purification and characterization of leukotriene A4 hydrolase from Xenopus laevis oocytes.'' FEBS Lett. 1998 Aug 21;433(3):219-22.]</ref> | |<ref name="Stromberg1998"> [www.ncbi.nlm.nih.gov/pubmed/9744798 Stromberg F. , "Purification and characterization of leukotriene A4 hydrolase from Xenopus laevis oocytes.'' FEBS Lett. 1998 Aug 21;433(3):219-22.]</ref> | ||
|- | |- | ||
| Line 48: | Line 56: | ||
pH: 8 | pH: 8 | ||
Temperature:2°C | Temperature:2°C | ||
| + | |256 | ||
|rowspan=2|<ref name="Radmark1984"> [www.ncbi.nlm.nih.gov/pubmed/6490615 Radmark O. , "Leukotriene A4 hydrolase in human leukocytes. Purification and properties'' J Biol Chem. 1984 Oct 25;259(20):12339-45.]</ref> | |rowspan=2|<ref name="Radmark1984"> [www.ncbi.nlm.nih.gov/pubmed/6490615 Radmark O. , "Leukotriene A4 hydrolase in human leukocytes. Purification and properties'' J Biol Chem. 1984 Oct 25;259(20):12339-45.]</ref> | ||
|- | |- | ||
| Line 55: | Line 64: | ||
pH: 8 | pH: 8 | ||
Temperature:37°C | Temperature:37°C | ||
| + | |1024 | ||
| + | | | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the LTA4H Kms distribution | ||
| + | ! Mode (mM) !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 2.40E-03 || 5.77E+00 || -5.51E+00 || 7.20E-01 | ||
| + | |} | ||
| + | |||
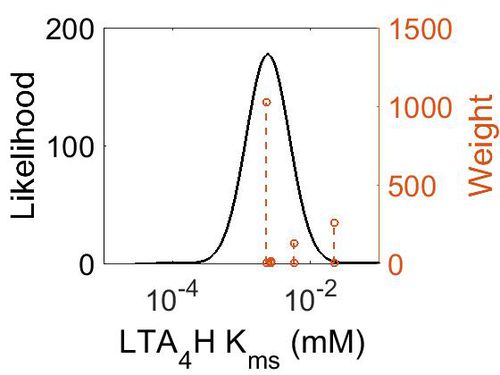
| + | [[Image:49.jpg|none|thumb|500px|The estimated probability distribution for LTA4H Kms. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
| + | |||
| + | |||
| + | ===K<sub>mp</sub>=== | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the LTA4H Kmp distribution | ||
| + | ! Mode (mM) !! Location parameter (µ) !! Scale parameter (σ) | ||
|- | |- | ||
| + | | 2.40E-03 || -5.51E+00 || 7.17E-01 | ||
|} | |} | ||
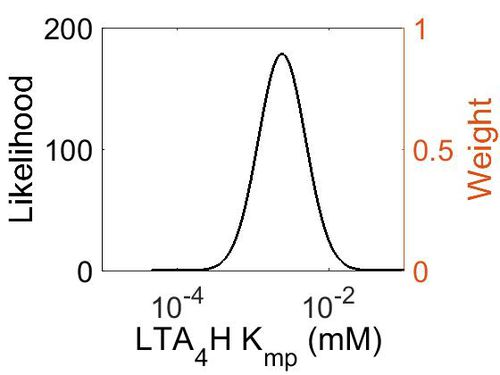
| + | [[Image:50.jpg|none|thumb|500px|The estimated probability distribution for LTA4H Kmp. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
| + | |||
| + | |||
| + | ===k<sub>cat</sub>=== | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Literature values |
|- | |- | ||
! Value | ! Value | ||
| Line 65: | Line 99: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 74: | Line 109: | ||
pH: 7.4 | pH: 7.4 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |1024 | ||
|<ref name="Rudberg2002"> [www.ncbi.nlm.nih.gov/pubmed/11675384 Rudberg P. , "Leukotriene A4 Hydrolase/Aminopeptidase'' The Journal of Biological Chemistry, 277, 1398-1404.]</ref> | |<ref name="Rudberg2002"> [www.ncbi.nlm.nih.gov/pubmed/11675384 Rudberg P. , "Leukotriene A4 Hydrolase/Aminopeptidase'' The Journal of Biological Chemistry, 277, 1398-1404.]</ref> | ||
|- | |- | ||
| Line 82: | Line 118: | ||
Enzyme: Wild Type Leukotriene A4 hydrolase: | Enzyme: Wild Type Leukotriene A4 hydrolase: | ||
pH:8 | pH:8 | ||
| − | Temperature: 20 (Room Temperature) | + | Temperature: 20 (Room Temperature) |
| + | |128 | ||
|<ref name="Mueller1996"> [www.ncbi.nlm.nih.gov/pubmed/8650196 Mueller M. , "Leukotriene A4 hydrolase: protection from mechanism-based inactivation by mutation of tyrosine-378.'' Proc Natl Acad Sci U S A. 1996 Jun 11;93(12):5931-5.]</ref> | |<ref name="Mueller1996"> [www.ncbi.nlm.nih.gov/pubmed/8650196 Mueller M. , "Leukotriene A4 hydrolase: protection from mechanism-based inactivation by mutation of tyrosine-378.'' Proc Natl Acad Sci U S A. 1996 Jun 11;93(12):5931-5.]</ref> | ||
|- | |- | ||
| Line 92: | Line 129: | ||
pH:8 | pH:8 | ||
Temperature: 20 (Room Temperature) | Temperature: 20 (Room Temperature) | ||
| + | |16 | ||
|<ref name="Stromberg1998"> [www.ncbi.nlm.nih.gov/pubmed/9744798 Stromberg F. , "Purification and characterization of leukotriene A4 hydrolase from Xenopus laevis oocytes.'' FEBS Lett. 1998 Aug 21;433(3):219-22.]</ref> | |<ref name="Stromberg1998"> [www.ncbi.nlm.nih.gov/pubmed/9744798 Stromberg F. , "Purification and characterization of leukotriene A4 hydrolase from Xenopus laevis oocytes.'' FEBS Lett. 1998 Aug 21;433(3):219-22.]</ref> | ||
|- | |- | ||
| Line 101: | Line 139: | ||
pH: 8 | pH: 8 | ||
Temperature:37°C | Temperature:37°C | ||
| + | |1024 | ||
|<ref name="Radmark1984"> [www.ncbi.nlm.nih.gov/pubmed/6490615 Radmark O. , "Leukotriene A4 hydrolase in human leukocytes. Purification and properties'' J Biol Chem. 1984 Oct 25;259(20):12339-45.]</ref> | |<ref name="Radmark1984"> [www.ncbi.nlm.nih.gov/pubmed/6490615 Radmark O. , "Leukotriene A4 hydrolase in human leukocytes. Purification and properties'' J Biol Chem. 1984 Oct 25;259(20):12339-45.]</ref> | ||
|- | |- | ||
|} | |} | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the LTA4H kcat distribution | ||
| + | ! Mode (min-1) !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 2.85E+01 || 6.56E+00 || 3.94E+00 || 7.60E-01 | ||
| + | |} | ||
| + | |||
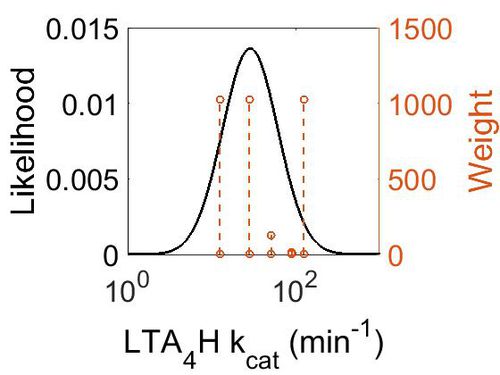
| + | [[Image:51.jpg|none|thumb|500px|The estimated probability distribution for LTA4H kcat. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
| + | |||
| + | |||
| + | ===Enzyme concentration=== | ||
| + | |||
| + | To convert the enzyme concentration from ppm to mM, the following [[Common equations#Enzyme concentration (mM)|equation]] was used. | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Literature values |
|- | |- | ||
! Value | ! Value | ||
| Line 112: | Line 165: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 121: | Line 175: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |1024 | ||
|<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | |<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | ||
|- | |- | ||
| Line 130: | Line 185: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |2048 | ||
|<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the human proteome'' Nature, 2014 509, 582–587]</ref> | |<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the human proteome'' Nature, 2014 509, 582–587]</ref> | ||
|- | |- | ||
| Line 139: | Line 195: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |1024 | ||
|<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the human proteome'' Nature, 2014 509, 582–587]</ref> | |<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the human proteome'' Nature, 2014 509, 582–587]</ref> | ||
|- | |- | ||
|} | |} | ||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the LTA4H concentration distribution | ||
| + | ! Mode (ppm) !! Mode (mM) !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 4.88E+02 ||2.70E-03|| 1.25E+00 || 6.24E+00 || 2.19E-01 | ||
| + | |} | ||
| + | |||
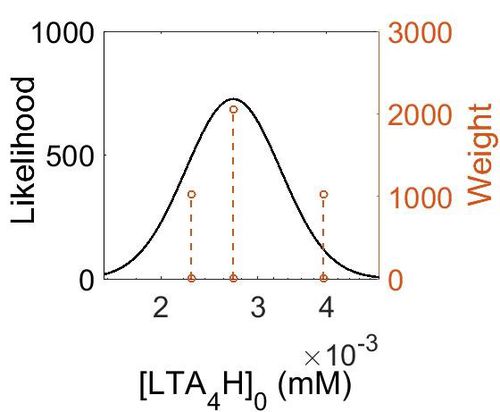
| + | [[Image:lta4h.jpg|none|thumb|500px|The estimated probability distribution for LTA4H concentration. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
| + | |||
| + | ===K<sub>eq</sub>=== | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Literature values |
|- | |- | ||
| − | ! | + | ! Gibbs Free Energy Change |
! Units | ! Units | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 161: | Line 229: | ||
pH: 7.3 | pH: 7.3 | ||
ionic strength: 0.25 | ionic strength: 0.25 | ||
| + | |64 | ||
|<ref name="MetaCyc”>[http://metacyc.org/META/NEW-IMAGE?type=REACTION&object=LEUKOTRIENE-A4-HYDROLASE-RXN Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471]</ref> | |<ref name="MetaCyc”>[http://metacyc.org/META/NEW-IMAGE?type=REACTION&object=LEUKOTRIENE-A4-HYDROLASE-RXN Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471]</ref> | ||
|} | |} | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the LTA4H Keq distribution | ||
| + | ! Mode !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 4.64E+02 || 1.00E+01 || 6.93E+00 || 8.90E-01 | ||
| + | |} | ||
| + | |||
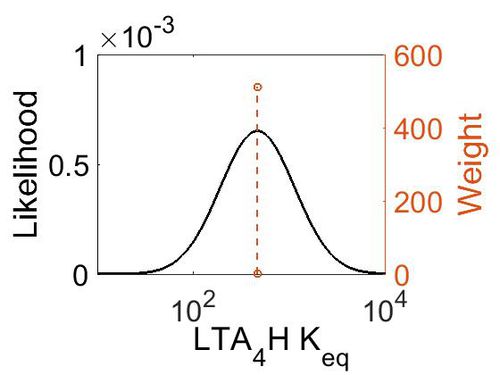
| + | [[Image:52.jpg|none|thumb|500px|The estimated probability distribution for LTA4H Keq. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
== References == | == References == | ||
Latest revision as of 08:39, 21 August 2019
LTA4 is subsequently enzymatically hydrolysed to LTB4 by LTA4 hydrolase (LTA4H). The enzyme performs this reaction by opening the epoxide and creating a carbocation intermediate. The charge of the carbocation delocalises over the triene system and is subsequently subject to nucleophilic attack by water at C12, resulting in the stereospecific addition of a hydroxyl group and the generation of LTB4.
Contents
Reaction
Chemical equation
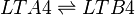
Rate equation
Parameters
Kms
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 5.80E-03 | mM | Human | Expression Vector: E. Coli
Enzyme: Wild Type Leukotriene A4 hydrolase: pH:8 Temperature: 20 (Room Temperature) |
128 | [1] |
| 2.70E-03 | mM | Frog | Expression Vector: oocytes
Enzyme: leukotriene A4 hydrolase pH:8 Temperature: 20 (Room Temperature) |
16 | [2] |
| 2.20E-02 | mM | Human | Expression Vector: Leukocytes
Enzyme: Leukotriene A4 Hydrolase pH: 8 Temperature:2°C |
256 | [3] |
| 2.30E-03 | Expression Vector: Leukocytes
Enzyme: Leukotriene A4 Hydrolase pH: 8 Temperature:37°C |
1024 |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 2.40E-03 | 5.77E+00 | -5.51E+00 | 7.20E-01 |
Kmp
| Mode (mM) | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 2.40E-03 | -5.51E+00 | 7.17E-01 |
kcat
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 12.6 - 28.2 | per minute | Human | Expression Vector: E. Coli.
Enzyme: Leukotriene A4 Hydrolase pH: 7.4 Temperature: 37 °C |
1024 | [4] |
| 51 | per minute | Human | Expression Vector: E. Coli
Enzyme: Wild Type Leukotriene A4 hydrolase: pH:8 Temperature: 20 (Room Temperature) |
128 | [1] |
| 90 | per minute | Frog | Expression Vector: oocytes
Enzyme: leukotriene A4 hydrolase pH:8 Temperature: 20 (Room Temperature) |
16 | [2] |
| 125 | per minute | Human | Expression Vector: Leukocytes
Enzyme: Leukotriene A4 Hydrolase pH: 8 Temperature:37°C |
1024 | [3] |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 2.85E+01 | 6.56E+00 | 3.94E+00 | 7.60E-01 |
Enzyme concentration
To convert the enzyme concentration from ppm to mM, the following equation was used.
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 714 | 
|
Human | Expression Vector: Lung
Enzyme: LTA4H pH: 7.5 Temperature: 37 °C |
1024 | [5] |
| 489 | 
|
Human | Expression Vector: Skin
Enzyme: LTA4H pH: 7.5 Temperature: 37 °C |
2048 | [6] |
| 410 | 
|
Human | Expression Vector: Oral Cavity
Enzyme: LTA4H pH: 7.5 Temperature: 37 °C |
1024 | [6] |
| Mode (ppm) | Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|---|
| 4.88E+02 | 2.70E-03 | 1.25E+00 | 6.24E+00 | 2.19E-01 |
Keq
| Gibbs Free Energy Change | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| (-3.6329994) | kcal/mol | Not stated | Estimated
Enzyme: LTA4H Substrate: LTA4 Product: LTB4 pH: 7.3 ionic strength: 0.25 |
64 | [7] |
| Mode | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 4.64E+02 | 1.00E+01 | 6.93E+00 | 8.90E-01 |
References
- ↑ 1.0 1.1 [www.ncbi.nlm.nih.gov/pubmed/8650196 Mueller M. , "Leukotriene A4 hydrolase: protection from mechanism-based inactivation by mutation of tyrosine-378. Proc Natl Acad Sci U S A. 1996 Jun 11;93(12):5931-5.]
- ↑ 2.0 2.1 [www.ncbi.nlm.nih.gov/pubmed/9744798 Stromberg F. , "Purification and characterization of leukotriene A4 hydrolase from Xenopus laevis oocytes. FEBS Lett. 1998 Aug 21;433(3):219-22.]
- ↑ 3.0 3.1 [www.ncbi.nlm.nih.gov/pubmed/6490615 Radmark O. , "Leukotriene A4 hydrolase in human leukocytes. Purification and properties J Biol Chem. 1984 Oct 25;259(20):12339-45.]
- ↑ [www.ncbi.nlm.nih.gov/pubmed/11675384 Rudberg P. , "Leukotriene A4 Hydrolase/Aminopeptidase The Journal of Biological Chemistry, 277, 1398-1404.]
- ↑ M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ 6.0 6.1 M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587
- ↑ Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471