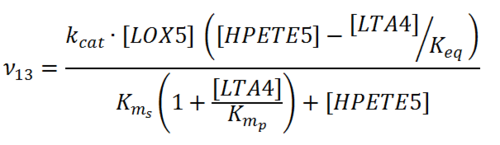Difference between revisions of "Transformation of 5-HPETE to LTA4"
(→Parameters) |
|||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Welcome to the In-Silico Model of Cutaneous Lipids Wiki | Return to overview]] | [[Welcome to the In-Silico Model of Cutaneous Lipids Wiki | Return to overview]] | ||
| − | The | + | In addition to being metabolised by PHGPx, 5-HPETE is also further metabolised by 5-LOX. This dehydration reaction converts the peroxide functional group of 5-HPETE to an epoxide functional group, generating LTA4 <ref>Shimizu, T. Radmark, O. Samuelsson, B., ''Enzyme with dual lipoxygenase activities catalyzes leukotriene A4 synthesis from arachidonic acid'', Proc Natl Acad Sci U S A (1984), 81, 689-93.</ref>. 5-LOX performs this reaction by abstracting the pro-R hydrogen at C10 and rearranging the structure so that the radical relocates to C6. The double bonds within the structure then rearrange to form a conjugated triene system and an epoxide. This reaction is promoted when 5-LOX colocalises with 5-lipoxygenase-activating protein (FLAP) on the nuclear membrane or the ER <ref>Abramovitz, M. Wong, E. Cox, M. E. Richardson, C. D. Li, C. Vickers, P. J. , ''5-lipoxygenase-activating protein stimulates the utilization of arachidonic acid by 5-lipoxygenase'', Eur J Biochem (1993), 215, 105-11.</ref><ref>Brock, T. G. Paine, R., 3rd Peters-Golden, M. , ''Localization of 5-lipoxygenase to the nucleus of unstimulated rat basophilic leukemia cells'', J Biol Chem (1994), 269, 22059-66.</ref>. |
| − | |||
== Reaction == | == Reaction == | ||
| Line 119: | Line 118: | ||
=== Enzyme concentration === | === Enzyme concentration === | ||
| + | |||
| + | To convert the enzyme concentration from ppm to mM, the following [[Common equations#Enzyme concentration (mM)|equation]] was used. | ||
| + | |||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
|+ style="text-align: left;" | Literature values | |+ style="text-align: left;" | Literature values | ||
| Line 163: | Line 165: | ||
{| class="wikitable" | {| class="wikitable" | ||
|+ style="text-align: left;" | Description of the 5-LOX concentration distribution | |+ style="text-align: left;" | Description of the 5-LOX concentration distribution | ||
| − | ! Mode (mM) !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | + | ! Mode (ppm) !! Mode (mM) !!Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) |
|- | |- | ||
| − | | 4.96E+01 || 1.60E+00 || 4.09E+00 || 4.28E-01 | + | | 4.96E+01 ||2.74E-04|| 1.60E+00 || 4.09E+00 || 4.28E-01 |
|} | |} | ||
[[Image:158.jpg|none|thumb|500px|The estimated probability distribution for 5-LOX/FLAP concentration. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | [[Image:158.jpg|none|thumb|500px|The estimated probability distribution for 5-LOX/FLAP concentration. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
| − | |||
=== K<sub>eq</sub> === | === K<sub>eq</sub> === | ||
Latest revision as of 08:36, 21 August 2019
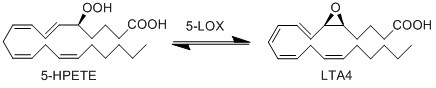
In addition to being metabolised by PHGPx, 5-HPETE is also further metabolised by 5-LOX. This dehydration reaction converts the peroxide functional group of 5-HPETE to an epoxide functional group, generating LTA4 [1]. 5-LOX performs this reaction by abstracting the pro-R hydrogen at C10 and rearranging the structure so that the radical relocates to C6. The double bonds within the structure then rearrange to form a conjugated triene system and an epoxide. This reaction is promoted when 5-LOX colocalises with 5-lipoxygenase-activating protein (FLAP) on the nuclear membrane or the ER [2][3].
Contents
Reaction
Chemical equation
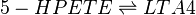
Rate equation
Parameters
Note that the literature values are the same as reaction 11.
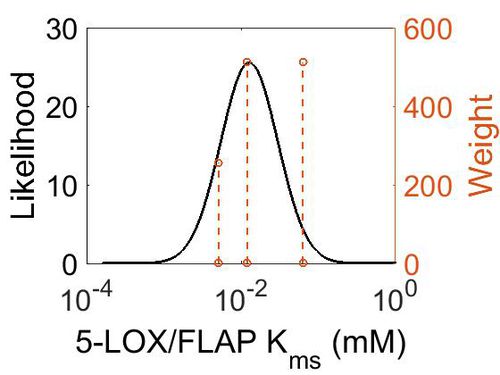
Kms
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 5.10E-03 | 
|
Human | Expression Vector: Baculovirus, Sf9 insect cells
Enzyme: Recombinant 5-Lipoxygenase pH: 5.6 Temperature:37 |
256 | [4] |
| 1.20E-02 | 
|
Human | Expression Vector: Polymorphonuclear Leukocytes
Enzyme: 5-Lipoxygenase pH:7.5 Temperature: 22 |
512 | [5] |
| 6.31E-02 | 
|
Human | Expression Vector: Polymorphonuclear Leukocytes
Enzyme: 5-Lipoxygenase pH:7.5 Temperature: 22 |
512 | [6] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.27E-02 | 2.74E+00 | -3.76E+00 | 7.73E-01 |
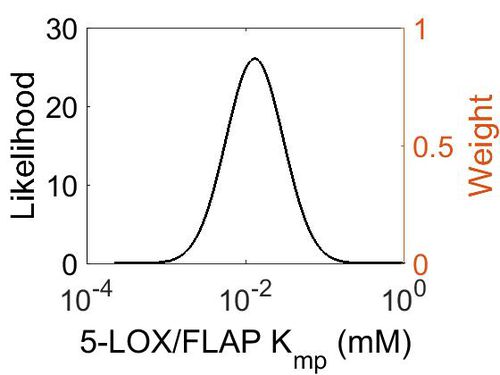
Kmp
| Mode (mM) | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 1.25E-02 | -3.63E+00 | 8.68E-01 |
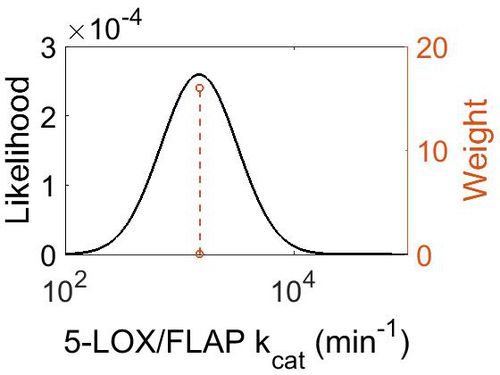
kcat
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 1500 + 75 | per minute | Potato | Expression Vector:Potato Tubers
Enzyme: 5-Lipoxygenase pH:5.5 Temperature: 23 |
16 | [7] |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.50E+03 | 1.05E+00 | 7.31E+00 | 4.99E-02 |
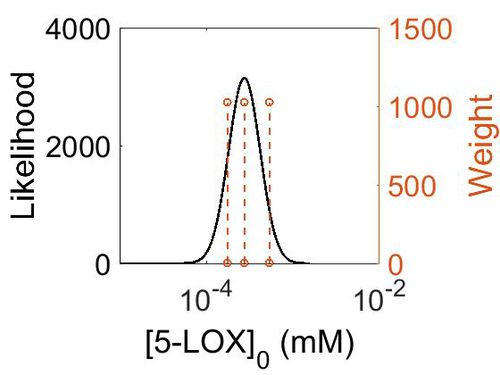
Enzyme concentration
To convert the enzyme concentration from ppm to mM, the following equation was used.
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 97.3 | 
|
Human | Expression Vector: Lung
Enzyme: 5-LOX pH: 7.5 Temperature: 37 °C |
1024 | [8] |
| 49.8 | 
|
Human | Expression Vector: Esophagus
Enzyme: 5-LOX pH: 7.5 Temperature: 37 °C |
1024 | [9] |
| 31.9 | 
|
Human | Expression Vector: Oral Cavity
Enzyme: 5-LOX pH: 7.5 Temperature: 37 °C |
1024 | [9] |
| Mode (ppm) | Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|---|
| 4.96E+01 | 2.74E-04 | 1.60E+00 | 4.09E+00 | 4.28E-01 |
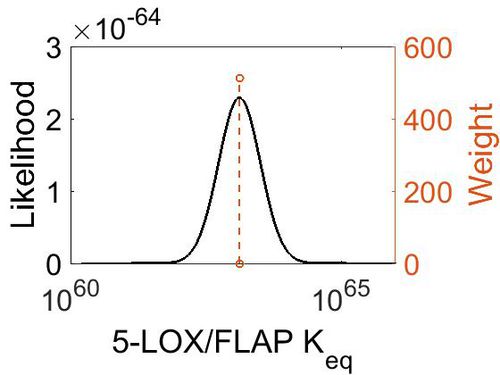
Keq
| Gibbs Free Energy Change | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| (-86.007) | kcal/mol | Not stated | Estimated
Enzyme: 5-LOX Substrate: 5-HPETE Product: LTA4 pH: 7.3 ionic strength: 0.25 |
64 | [10] |
| Mode | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.31E+63 | 1.00E+01 | 1.46E+02 | 8.90E-01 |
References
- ↑ Shimizu, T. Radmark, O. Samuelsson, B., Enzyme with dual lipoxygenase activities catalyzes leukotriene A4 synthesis from arachidonic acid, Proc Natl Acad Sci U S A (1984), 81, 689-93.
- ↑ Abramovitz, M. Wong, E. Cox, M. E. Richardson, C. D. Li, C. Vickers, P. J. , 5-lipoxygenase-activating protein stimulates the utilization of arachidonic acid by 5-lipoxygenase, Eur J Biochem (1993), 215, 105-11.
- ↑ Brock, T. G. Paine, R., 3rd Peters-Golden, M. , Localization of 5-lipoxygenase to the nucleus of unstimulated rat basophilic leukemia cells, J Biol Chem (1994), 269, 22059-66.
- ↑ Shirumalla R. K. “RBx 7,796: A novel inhibitor of 5-lipoxygenase.” Inflamm Res. 2006 Dec ; 55 (12) : 517-27.
- ↑ Soberman R. J. "5- and 15(omega-6)-lipoxygenases from human polymorphonuclear leukocytes. Methods Enzymol. 1988; 163:344-9.
- ↑ Soberman R. J. “Characterization and separation of the arachidonic acid 5-lipoxygenase and linoleic acid omega-6 lipoxygenase (arachidonic acid 15-lipoxygenase) of human polymorphonuclear leukocytes.” J Biol Chem. 1985 Apr 10;260(7):4508-15.
- ↑ Mulliez E., “5-Lipoxygenase from potato tubers. Improved purification and physicochemical characteristics” Biochimica et Biophysica Acta, 1987;916(1):13-23.
- ↑ M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ 9.0 9.1 M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587
- ↑ Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471