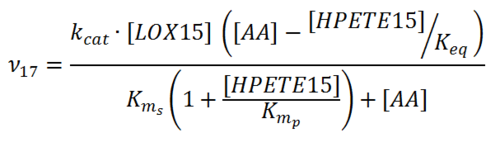Difference between revisions of "Transformation of AA to 15-HPETE"
| (4 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Welcome to the In-Silico Model of Cutaneous Lipids Wiki | Return to overview]] | [[Welcome to the In-Silico Model of Cutaneous Lipids Wiki | Return to overview]] | ||
| − | + | The genes ALOX15B and ALOX15 encodes the formation of two isoforms of 15-lioxygenase (15-LOX). The gene ALOX15B encodes the formation of 15-LOX-2, which converts AA exclusively into 15(S)-HPETE. Whereas the gene ALOX15 encodes the 15-LOX-1 protein, which converts AA into both 12(S)-HPETE and 15(S)-HPETE. The formation of the unstable hydroperoxy fatty acids (HPETE) begins with the abstraction of a hydrogen radical at the allylic position between two double bonds. The structure undergoes a rearrangement reaction which results in the formation of a conjugated diene system. The insertion of molecular oxygen and a hydrogen leads to the formation of the final structure, a hydroperoxy fatty acid. | |
| − | |||
== Reaction == | == Reaction == | ||
| Line 16: | Line 15: | ||
== 15-LOX Parameters == | == 15-LOX Parameters == | ||
| − | + | ===K<sub>ms</sub>=== | |
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Literature values |
|- | |- | ||
! Value | ! Value | ||
| Line 24: | Line 23: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 33: | Line 33: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 25 | Temperature: 25 | ||
| + | |512 | ||
|<ref name="Jacquot2008"> [http://www.ncbi.nlm.nih.gov/pubmed/18547056 Jacquot C. "Isotope sensitive branching and kinetic isotope effects in the reaction of deuterated arachidonic acids with human 12- and 15-lipoxygenases.'' Biochemistry. 2008 Jul 8;47(27):7295-303. doi: 10.1021/bi800308q. Epub 2008 Jun 12.]</ref> | |<ref name="Jacquot2008"> [http://www.ncbi.nlm.nih.gov/pubmed/18547056 Jacquot C. "Isotope sensitive branching and kinetic isotope effects in the reaction of deuterated arachidonic acids with human 12- and 15-lipoxygenases.'' Biochemistry. 2008 Jul 8;47(27):7295-303. doi: 10.1021/bi800308q. Epub 2008 Jun 12.]</ref> | ||
|- | |- | ||
| Line 42: | Line 43: | ||
pH:7.5 | pH:7.5 | ||
Temperature: Unspecified | Temperature: Unspecified | ||
| + | |512 | ||
|<ref name="Jacquot2008b"> [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2883171/ Jacquot C. "Synthesis of 11-Thialinoleic Acid and 14-Thialinoleic Acid, Inhibitors of Soybean and Human Lipoxygenases'' Org Biomol Chem. 2008 Nov 21; 6(22): 4242–4252.]</ref> | |<ref name="Jacquot2008b"> [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2883171/ Jacquot C. "Synthesis of 11-Thialinoleic Acid and 14-Thialinoleic Acid, Inhibitors of Soybean and Human Lipoxygenases'' Org Biomol Chem. 2008 Nov 21; 6(22): 4242–4252.]</ref> | ||
|- | |- | ||
| Line 51: | Line 53: | ||
pH: 6.7 - 7.3 | pH: 6.7 - 7.3 | ||
Temperature: 37 | Temperature: 37 | ||
| + | |2048 | ||
|<ref name="Burrall1988"> [http://www.ncbi.nlm.nih.gov/pubmed/2459258 Burrall B. "Enzymatic properties of the 15-lipoxygenase of human cultured keratinocytes.'' J Invest Dermatol. 1988 Oct;91(4):294-7.]</ref> | |<ref name="Burrall1988"> [http://www.ncbi.nlm.nih.gov/pubmed/2459258 Burrall B. "Enzymatic properties of the 15-lipoxygenase of human cultured keratinocytes.'' J Invest Dermatol. 1988 Oct;91(4):294-7.]</ref> | ||
|- | |- | ||
| Line 60: | Line 63: | ||
pH: 6.8 | pH: 6.8 | ||
Temperature: 37 | Temperature: 37 | ||
| + | |512 | ||
|<ref name="Sloane1995"> [http://www.ncbi.nlm.nih.gov/pubmed/7479689 Sloane D. L. "Conversion of human 15-lipoxygenase to an efficient 12-lipoxygenase: the side-chain geometry of amino acids 417 and 418 determine positional specificity.'' Protein Eng. 1995 Mar;8(3):275-82..]</ref> | |<ref name="Sloane1995"> [http://www.ncbi.nlm.nih.gov/pubmed/7479689 Sloane D. L. "Conversion of human 15-lipoxygenase to an efficient 12-lipoxygenase: the side-chain geometry of amino acids 417 and 418 determine positional specificity.'' Protein Eng. 1995 Mar;8(3):275-82..]</ref> | ||
|- | |- | ||
|} | |} | ||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the 15-LOX Kms distribution | ||
| + | ! Mode (mM) !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 1.02E-02 || 2.40E+00 || -4.42E+00 || 4.10E-01 | ||
| + | |- | ||
| + | |} | ||
| + | |||
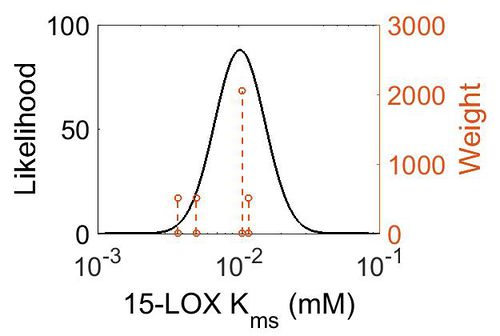
| + | [[Image:57.jpg|none|thumb|500px|The estimated probability distribution for 15-LOX Kms. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
| + | |||
| + | |||
| + | ===K<sub>mp</sub>=== | ||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the 15-LOX Kmp distribution | ||
| + | ! Mode (mM) !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 1.02E-02 || -4.42E+00 || 4.11E-01 | ||
| + | |} | ||
| + | |||
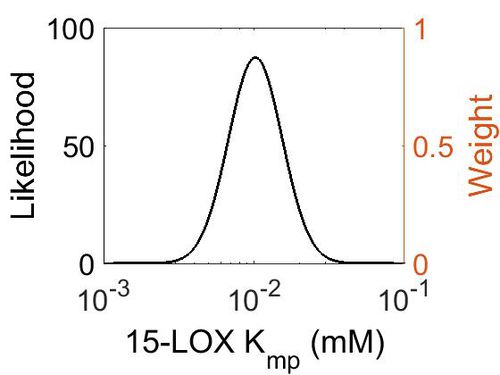
| + | [[Image:58.jpg|none|thumb|500px|The estimated probability distribution for 15-LOX Kmp. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
| + | |||
| + | ===k<sub>cat</sub>=== | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Literature values |
|- | |- | ||
! Value | ! Value | ||
| Line 71: | Line 97: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 80: | Line 107: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 25 | Temperature: 25 | ||
| + | |512 | ||
|<ref name="Jacquot2008"> [http://www.ncbi.nlm.nih.gov/pubmed/18547056 Jacquot C. "Isotope sensitive branching and kinetic isotope effects in the reaction of deuterated arachidonic acids with human 12- and 15-lipoxygenases.'' Biochemistry. 2008 Jul 8;47(27):7295-303. doi: 10.1021/bi800308q. Epub 2008 Jun 12.]</ref> | |<ref name="Jacquot2008"> [http://www.ncbi.nlm.nih.gov/pubmed/18547056 Jacquot C. "Isotope sensitive branching and kinetic isotope effects in the reaction of deuterated arachidonic acids with human 12- and 15-lipoxygenases.'' Biochemistry. 2008 Jul 8;47(27):7295-303. doi: 10.1021/bi800308q. Epub 2008 Jun 12.]</ref> | ||
|- | |- | ||
| Line 87: | Line 115: | ||
Enzyme:15-lipoxygenase-2 | Enzyme:15-lipoxygenase-2 | ||
|pH 7, Temperature: 22 | |pH 7, Temperature: 22 | ||
| + | |rowspan="7"|512 | ||
|rowspan="7"|<ref name="Wecksler2009"> [http://www.ncbi.nlm.nih.gov/pubmed/18547056 Wecksler A. "Kinetic and Structural Investigations of the Allosteric Site in Human Epithelial 15-Lipoxygenase-2'' Biochemistry, 2009, 48 (36), pp 8721–8730]</ref> | |rowspan="7"|<ref name="Wecksler2009"> [http://www.ncbi.nlm.nih.gov/pubmed/18547056 Wecksler A. "Kinetic and Structural Investigations of the Allosteric Site in Human Epithelial 15-Lipoxygenase-2'' Biochemistry, 2009, 48 (36), pp 8721–8730]</ref> | ||
|- | |- | ||
| Line 108: | Line 137: | ||
|- | |- | ||
|} | |} | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the 15-LOX kcat distribution | ||
| + | ! Mode (min-1) !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 6.51E+01 || 4.62E+00 || 4.60E+00 || 6.50E-01 | ||
| + | |} | ||
| + | |||
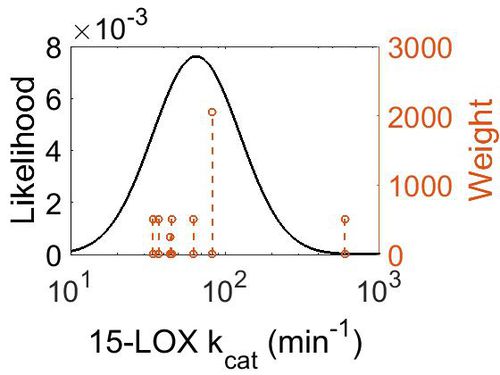
| + | [[Image:59.jpg|none|thumb|500px|The estimated probability distribution for 15-LOX kcat. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
| + | |||
| + | ===Enzyme concentration === | ||
| + | |||
| + | To convert the enzyme concentration from ppm to mM, the following [[Common equations#Enzyme concentration (mM)|equation]] was used. | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Literature values |
|- | |- | ||
! Value | ! Value | ||
| Line 116: | Line 158: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 125: | Line 168: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |1024 | ||
|<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | |<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | ||
|- | |- | ||
| Line 134: | Line 178: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |1024 | ||
|<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the human proteome'' Nature, 2014 509, 582–587]</ref> | |<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the human proteome'' Nature, 2014 509, 582–587]</ref> | ||
|- | |- | ||
| Line 143: | Line 188: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |1024 | ||
|<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | |<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | ||
|- | |- | ||
|} | |} | ||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the 15-LOX concentration distribution | ||
| + | ! Mode (ppm) !! Mode (mM) !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 4.07 || 2.25E-05|| 7.23E+00 || 7.12E-01 || 1.19E+00 | ||
| + | |} | ||
| + | |||
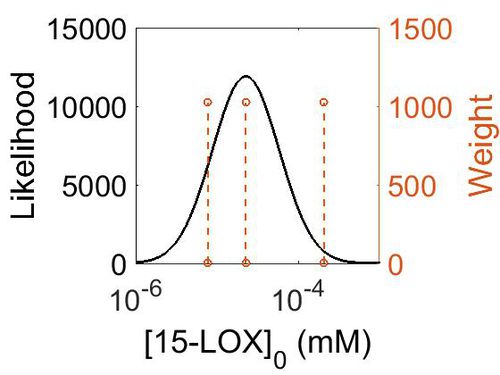
| + | [[Image:157.jpg|none|thumb|500px|The estimated probability distribution for 15-LOX concentration. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
| + | |||
| + | ===K<sub>eq</sub>=== | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Literature values |
|- | |- | ||
| − | ! | + | ! Gibbs Free Energy Change |
! Units | ! Units | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 165: | Line 222: | ||
pH: 7.3 | pH: 7.3 | ||
ionic strength: 0.25 | ionic strength: 0.25 | ||
| + | |64 | ||
|<ref name="MetaCyc”>[http://metacyc.org/META/NEW-IMAGE?type=REACTION&object=RXN66-490 Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471]</ref> | |<ref name="MetaCyc”>[http://metacyc.org/META/NEW-IMAGE?type=REACTION&object=RXN66-490 Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471]</ref> | ||
|} | |} | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the 15-LOX Keq distribution | ||
| + | ! Mode !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 2.27E+51 || 1.00E+01 || 1.19E+02 || 8.90E-01 | ||
| + | |} | ||
| + | |||
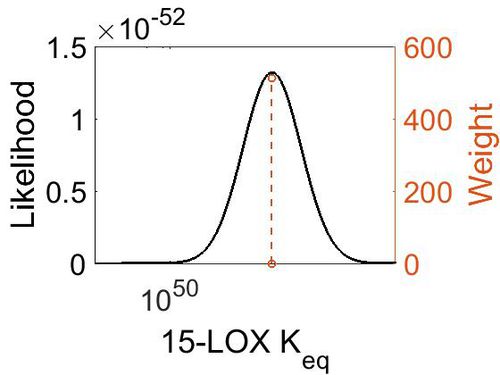
| + | [[Image:60.jpg|none|thumb|500px|The estimated probability distribution for 15-LOX Keq. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
== References == | == References == | ||
Latest revision as of 08:19, 21 August 2019
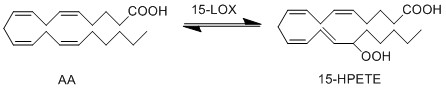
The genes ALOX15B and ALOX15 encodes the formation of two isoforms of 15-lioxygenase (15-LOX). The gene ALOX15B encodes the formation of 15-LOX-2, which converts AA exclusively into 15(S)-HPETE. Whereas the gene ALOX15 encodes the 15-LOX-1 protein, which converts AA into both 12(S)-HPETE and 15(S)-HPETE. The formation of the unstable hydroperoxy fatty acids (HPETE) begins with the abstraction of a hydrogen radical at the allylic position between two double bonds. The structure undergoes a rearrangement reaction which results in the formation of a conjugated diene system. The insertion of molecular oxygen and a hydrogen leads to the formation of the final structure, a hydroperoxy fatty acid.
Contents
Reaction
Chemical equation
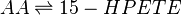
Rate equation
15-LOX Parameters
Kms
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 3.70E-03 ± 3.00E-04 | 
|
Human | Expression Vector: Epithelium
Enzyme: 15-Lipoxygenase pH: 7.5 Temperature: 25 |
512 | [1] |
| 5.00E-03 | 
|
Human | Expression Vector: Reticulocyte
Enzyme: 15-Lipoxygenase pH:7.5 Temperature: Unspecified |
512 | [2] |
| 1.06E-02 | 
|
Human | Expression Vector: Keratinocyte
Enzyme: 15-Lipoxygenase pH: 6.7 - 7.3 Temperature: 37 |
2048 | [3] |
| 1.17E-02 ± 9.00E-04 | 
|
Human | Expression Vector: Baculovirus Insect Cell
Enzyme: Wildtype 15-Lipoxygenase pH: 6.8 Temperature: 37 |
512 | [4] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.02E-02 | 2.40E+00 | -4.42E+00 | 4.10E-01 |
Kmp
| Mode (mM) | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 1.02E-02 | -4.42E+00 | 4.11E-01 |
kcat
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 595.8 ± 16.8 | 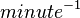
|
Human | Expression Vector: Epithelium
Enzyme: 15-Lipoxygenase pH: 7.5 Temperature: 25 |
512 | [1] |
| 37.2 ± 1.8 | per minute | Expression Vector:Human prostate epithelial
Enzyme:15-lipoxygenase-2 |
pH 7, Temperature: 22 | 512 | [5] |
| 45 ± 1.2 | pH 7.5, Temperature: 22 | ||||
| 44.4 ± 2.4 | pH 8, Temperature: 22 | ||||
| 34.2 ± 0.6 | 15°C, pH = 7.5 | ||||
| 45 ± 1.2 | 22°C, pH = 7.5 | ||||
| 62.4 ± 4.2 | 30°C, pH = 7.5 | ||||
| 82.8 ± 4.8 | 37°C, pH = 7.5 |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 6.51E+01 | 4.62E+00 | 4.60E+00 | 6.50E-01 |
Enzyme concentration
To convert the enzyme concentration from ppm to mM, the following equation was used.
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 4.09 | 
|
Human | Expression Vector: Lung
Enzyme: 15-LOX pH: 7.5 Temperature: 37 °C |
1024 | [6] |
| 37.4 | 
|
Human | Expression Vector: Spleen
Enzyme: 15-LOX pH: 7.5 Temperature: 37 °C |
1024 | [7] |
| 1.40 | 
|
Human | Expression Vector: Gut
Enzyme: 12-LOX pH: 7.5 Temperature: 37 °C |
1024 | [6] |
| Mode (ppm) | Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|---|
| 4.07 | 2.25E-05 | 7.23E+00 | 7.12E-01 | 1.19E+00 |
Keq
| Gibbs Free Energy Change | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| (-69.979996) | kcal/mol | Not stated | Estimated
Enzyme: 15-LOX Substrate: Arachidonate Product: 15-HPETE pH: 7.3 ionic strength: 0.25 |
64 | [8] |
| Mode | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 2.27E+51 | 1.00E+01 | 1.19E+02 | 8.90E-01 |
References
- ↑ 1.0 1.1 Jacquot C. "Isotope sensitive branching and kinetic isotope effects in the reaction of deuterated arachidonic acids with human 12- and 15-lipoxygenases. Biochemistry. 2008 Jul 8;47(27):7295-303. doi: 10.1021/bi800308q. Epub 2008 Jun 12.
- ↑ Jacquot C. "Synthesis of 11-Thialinoleic Acid and 14-Thialinoleic Acid, Inhibitors of Soybean and Human Lipoxygenases Org Biomol Chem. 2008 Nov 21; 6(22): 4242–4252.
- ↑ Burrall B. "Enzymatic properties of the 15-lipoxygenase of human cultured keratinocytes. J Invest Dermatol. 1988 Oct;91(4):294-7.
- ↑ Sloane D. L. "Conversion of human 15-lipoxygenase to an efficient 12-lipoxygenase: the side-chain geometry of amino acids 417 and 418 determine positional specificity. Protein Eng. 1995 Mar;8(3):275-82..
- ↑ Wecksler A. "Kinetic and Structural Investigations of the Allosteric Site in Human Epithelial 15-Lipoxygenase-2 Biochemistry, 2009, 48 (36), pp 8721–8730
- ↑ 6.0 6.1 M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587
- ↑ Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471