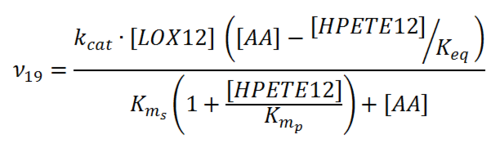Difference between revisions of "Transformation of AA to 12-HPETE"
(→12-LOX Parameters) |
|||
| Line 15: | Line 15: | ||
[[File:R19.PNG|center|500px]] | [[File:R19.PNG|center|500px]] | ||
| − | == | + | == Enzyme Parameters == |
| − | + | === K<sub>ms</sub>=== | |
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Literature values |
|- | |- | ||
! Value | ! Value | ||
| Line 64: | Line 64: | ||
|} | |} | ||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the 12-LOX Kms distribution | ||
| + | ! Mode (mM) !! Confidence Interval !! Location parameter (σ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 7.60E-03 || 4.34E+00 || -4.48E+00 || 6.30E-01 | ||
| + | |} | ||
| + | |||
| + | |||
| + | ===K<sub>mp</sub>=== | ||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the 12-LOX Kmp distribution | ||
| + | ! Mode (mM) !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 7.30E-03 || -4.52E+00 || 6.28E-01 | ||
| + | |} | ||
| + | |||
| + | ===k<sub>cat</sub>=== | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Literature values |
|- | |- | ||
! Value | ! Value | ||
| Line 91: | Line 108: | ||
|<ref name="Richards1997"> [http://pubs.acs.org/doi/abs/10.1021/bi963051a Richards K. "Leukocyte 12-Lipoxygenase: Expression, Purification, and Investigation of the Role of Methionine Residues in Turnover-Dependent Inactivation and 5,8,11,14-Eicosatetraynoic Acid Inhibition'' Biochemistry, 1997, 36 (22), pp 6692–6699]</ref> | |<ref name="Richards1997"> [http://pubs.acs.org/doi/abs/10.1021/bi963051a Richards K. "Leukocyte 12-Lipoxygenase: Expression, Purification, and Investigation of the Role of Methionine Residues in Turnover-Dependent Inactivation and 5,8,11,14-Eicosatetraynoic Acid Inhibition'' Biochemistry, 1997, 36 (22), pp 6692–6699]</ref> | ||
|- | |- | ||
| + | |} | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the 12-LOX kcat distribution | ||
| + | ! Mode (min-1) !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 4.87E+02 || 1.20E+00 || 6.22E+00 || 1.80E-01 | ||
|} | |} | ||
| + | === Enzyme concentration === | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
|+ style="text-align: left;" | 12-LOX Abundance | |+ style="text-align: left;" | 12-LOX Abundance | ||
| Line 141: | Line 166: | ||
|} | |} | ||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the 12-LOX concentration distribution | ||
| + | ! Mode (mM) !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 4.98E-01 || 7.23E+00 || 7.12E-01 || 1.19E+00 | ||
| + | |} | ||
| + | |||
| + | ===K<sub>eq</sub>=== | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Literature values |
|- | |- | ||
| − | ! | + | ! Gibbs Free Energy Change |
! Units | ! Units | ||
! Species | ! Species | ||
| Line 160: | Line 193: | ||
ionic strength: 0.25 | ionic strength: 0.25 | ||
|<ref name="MetaCyc”>[http://metacyc.org/META/NEW-IMAGE?type=REACTION&object=ARACHIDONATE-12-LIPOXYGENASE-RXN Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471]</ref> | |<ref name="MetaCyc”>[http://metacyc.org/META/NEW-IMAGE?type=REACTION&object=ARACHIDONATE-12-LIPOXYGENASE-RXN Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471]</ref> | ||
| + | |} | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |+ style="text-align: left;" | Description of the 12-LOX Keq distribution | ||
| + | ! Mode !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ||
| + | |- | ||
| + | | 2.27E+51 || 1.00E+01 || 1.19E+02 || 8.90E-01 | ||
|} | |} | ||
Revision as of 06:46, 16 May 2019
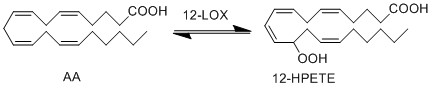
LOX enzymes oxidise AA to generate hydroxy fatty acids. In the skin, the 12-LOX isoform of LOX is more active than others. This isoform is highly expressed in the resident cells of both compartments, epidermal keratinocytes and dermal fibroblasts (Dowd, Kobza Black et al. 1985). The 12- LOX enzyme catalyses the addition of O2 at the C-12 position of AA, producing 12-HPETE.
Contents
Reaction
Chemical equation
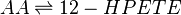
Rate equation
Enzyme Parameters
Kms
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 7.20E-03 | 
|
Human | Expression Vector: Platelet
Enzyme: 12-Lipoxygenase pH: 7.4 Temperature: 37°C. |
[1] |
| 8.00E-02 | 
|
Human | Expression Vector: Platelet
Enzyme: 12-Lipoxygenase pH: 7 Temperature: 24 |
[2] |
| 1.00E-02 | 
|
Human Platlet | Expression Vector: Baculovirus
Enzyme: 12-Lipoxygenase pH: 8 Temperature: 37 |
[3] |
| 7.90E-03 ± 8.00E-04 | 
|
Human | Expression Vector: Platelets
Enzyme:12-Lipoxygenase pH: 7.4 Temperature: 37 |
[4] |
| Mode (mM) | Confidence Interval | Location parameter (σ) | Scale parameter (σ) |
|---|---|---|---|
| 7.60E-03 | 4.34E+00 | -4.48E+00 | 6.30E-01 |
Kmp
| Mode (mM) | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 7.30E-03 | -4.52E+00 | 6.28E-01 |
kcat
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 336 ± 12 | per minute | Human | Expression Vector: Human reticulocyte
Enzyme: 15-lipoxygenase-1 pH: 7.5 Temperature: 25 |
[5] |
| 504 | per minute | Wild Boar | Expression Vector: E. coli.
Enzyme: 12-Lipoxygenase pH: 7.4 Temperature: 37 |
[6] |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 4.87E+02 | 1.20E+00 | 6.22E+00 | 1.80E-01 |
Enzyme concentration
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 19.8 | 
|
Human | Expression Vector: Spleen
Enzyme: 12-LOX pH: 7.5 Temperature: 37 °C |
[7] |
| 1.60 | 
|
Human | Expression Vector: Liver
Enzyme: 12-LOX pH: 7.5 Temperature: 37 °C |
[8] |
| 0.28 | 
|
Human | Expression Vector: Gut
Enzyme: 12-LOX pH: 7.5 Temperature: 37 °C |
[8] |
| 0.11 | 
|
Human | Expression Vector: Pancreas
Enzyme: 12-LOX pH: 7.5 Temperature: 37 °C |
[8] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 4.98E-01 | 7.23E+00 | 7.12E-01 | 1.19E+00 |
Keq
| Gibbs Free Energy Change | Units | Species | Notes | Reference |
|---|---|---|---|---|
| (-69.979996) | kcal/mol | Not stated | Estimated
Enzyme: 12-LOX Substrate: Arachidonate Product: 12-HPETE pH: 7.3 ionic strength: 0.25 |
[9] |
| Mode | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 2.27E+51 | 1.00E+01 | 1.19E+02 | 8.90E-01 |
References
- ↑ Lagarde M. "Subcellular localization and some properties of lipoxygenase activity in human blood platelets. Biochem J. 1984 Sep 1;222(2):495-500.
- ↑ Hada T. "Catalytic properties of human platelet 12-lipoxygenase as compared with the enzymes of other origins. Biochim Biophys Acta. 1991 Apr 24;1083(1):89-93.
- ↑ Chen X. S. "Purification and characterization of recombinant histidine-tagged human platelet 12-lipoxygenase expressed in a baculovirus/insect cell system. Eur J Biochem. 1993 Jun 15;214(3):845-52.
- ↑ Romano M. "Lipoxin synthase activity of human platelet 12-lipoxygenase. Biochem J. 1993 Nov 15;296 ( Pt 1):127-33.
- ↑ [www.ncbi.nlm.nih.gov/pubmed/19469483 Wecksler A. "Mechanistic Investigations of Human Reticulocyte 15- and Platelet 12-Lipoxygenases with Arachidonic Acid Biochemistry, 2009, 48 (26), pp 6259–6267]
- ↑ Richards K. "Leukocyte 12-Lipoxygenase: Expression, Purification, and Investigation of the Role of Methionine Residues in Turnover-Dependent Inactivation and 5,8,11,14-Eicosatetraynoic Acid Inhibition Biochemistry, 1997, 36 (22), pp 6692–6699
- ↑ M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587
- ↑ 8.0 8.1 8.2 M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471