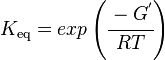Difference between revisions of "Double-bond reductase (DBR)"
Aliah.hawari (talk | contribs) (→Equation Rate) |
Aliah.hawari (talk | contribs) (→Reaction catalysed) |
||
| (18 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
You can go back to main page of the kinetic model [http://www.systemsbiology.ls.manchester.ac.uk/wiki/index.php/Kinetic_Model_of_Monoterpenoid_Biosynthesis_Wiki here]. | You can go back to main page of the kinetic model [http://www.systemsbiology.ls.manchester.ac.uk/wiki/index.php/Kinetic_Model_of_Monoterpenoid_Biosynthesis_Wiki here]. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== Reaction catalysed == | == Reaction catalysed == | ||
| − | + | [[File:DBR_reaction_diagram_v2.png | left | 500px ]] | |
| − | : | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== Enzyme and Metabolite Background Information == | == Enzyme and Metabolite Background Information == | ||
| Line 49: | Line 12: | ||
{| class="wikitable" style="text-align:center" | {| class="wikitable" style="text-align:center" | ||
|- | |- | ||
| − | ! style="border: 1px solid black; padding: 5px; background: #ADD8E6;"| | + | ! style="border: 1px solid black; padding: 5px; background: #ADD8E6;"|Compound |
! style="border: 1px solid black; padding: 5px; background: #ADD8E6;"|Abbreviation | ! style="border: 1px solid black; padding: 5px; background: #ADD8E6;"|Abbreviation | ||
! style="border: 1px solid black; padding: 5px; background: #ADD8E6;"|Chemical Formula | ! style="border: 1px solid black; padding: 5px; background: #ADD8E6;"|Chemical Formula | ||
| Line 59: | Line 22: | ||
! style="border: 1px solid black; padding: 5px; background: #ADD8E6;"|PlantCyc | ! style="border: 1px solid black; padding: 5px; background: #ADD8E6;"|PlantCyc | ||
|- | |- | ||
| − | | | + | | Double-bond reductase |
| − | | | + | | DBR |
| | | | ||
| 37914 Da | | 37914 Da | ||
| Line 123: | Line 86: | ||
== Equation Rate == | == Equation Rate == | ||
| − | + | DBR is modelled using two equations that describes: 1) Pulegone to Menthone and 2) Pulegone to Isomenthone. The predominant product for DBR is menthone where the ratio of menthone to isomenthone production have been reported as 40:33 <ref name="Toogood2015"> [http://www.ncbi.nlm.nih.gov/pubmed/26017480 Toogood, H. ''et. al.''2015. "Enzymatic menthol production: One-pot approach using engioneered ''Escherichia coli'' "], ACS Synthetic Biology, '''4''':1112-1123</ref>, 55:45 <ref name="Ringer2003">[http://www.ncbi.nlm.nih.gov/pubmed/13679086 Ringer, K.L. ''et. al.'' 2003. "Monoterpene double-bond reductases of the (-)-menthol biosynthetic pathway:isolation and characterization of cDNAs encoding (-)-isopiperitenone reductase and (+)-pulegone reductase of peppermint"], Archives of Biochemistry and Biophysics '''418''':80-92</ref> and 70:30 <ref name="Solodar1976">[http://pubs.acs.org/doi/abs/10.1021/jo00883a031 Solodar, J. 1976. "Stereoselective reduction of menthone and isomenthone by dissolving alkali metals and by hydrogen with Group 8 Metals on carbon"], The Journal of Organic Chemistry, '''41'''(21):3461-3464</ref>. To model the variability of the product yield for DBR, variability factors which are calculated from the average ratio is included in the equations. | |
| + | |||
| + | <ref name="Toogood2015"> [http://www.ncbi.nlm.nih.gov/pubmed/26017480 Toogood, H. ''et. al.'']2015. "Enzymatic menthol production: One-pot approach using engioneered ''Escherichia coli'' ", ACS Synthetic Biology, '''4''':1112-1123</ref> | ||
''' Reaction 1: Conversion of pulegone to menthone''' | ''' Reaction 1: Conversion of pulegone to menthone''' | ||
:<math> | :<math> | ||
| − | V_\mathrm{ | + | V_\mathrm{DBR} = Kcat_\mathrm{forward} * X1*[DBR] * \cfrac {\left ( \cfrac{[pulegone]}{Km_\mathrm{pulegone}} * \cfrac {[NADPH]}{Km_\mathrm{NADPH}} \right ) * \left ( 1 - \cfrac {[menthone]*[NADP]}{[pulegone]*[NADPH]*K_\mathrm{eq}} \right )} |
| − | { \left (1 + \cfrac {[ | + | { \left (1 + \cfrac {[Pulegone]}{Km_\mathrm{Pulegone}} + \cfrac {[NADP]}{Km_\mathrm{NADP}} \right ) + \left ( 1+ \cfrac {[NADPH]}{Km_\mathrm{NADPH}} + \cfrac {[menthone]}{Km_\mathrm{menthone}} \right ) } |
</math> | </math> | ||
''' Reaction 2: Conversion of pulegone to isomenthone''' | ''' Reaction 2: Conversion of pulegone to isomenthone''' | ||
:<math> | :<math> | ||
| − | V_\mathrm{ | + | V_\mathrm{DBR} = Kcat_\mathrm{forward} *X2* [DBR] * \cfrac {\left ( \cfrac{[pulegone]}{Km_\mathrm{pulegone}} * \cfrac {[NADPH]}{Km_\mathrm{NADPH}} \right ) * \left ( 1 - \cfrac {[isomenthone]*[NADP]}{[pulegone]*[NADPH]*K_\mathrm{eq}} \right )} |
| − | { \left (1 + \cfrac {[ | + | { \left (1 + \cfrac {[Pulegone]}{Km_\mathrm{Pulegone}} + \cfrac {[NADP]}{Km_\mathrm{NADP}} \right ) + \left ( 1+ \cfrac {[NADPH]}{Km_\mathrm{NADPH}} + \cfrac {[isomenthone]}{Km_\mathrm{isomenthone}} \right ) } |
</math> | </math> | ||
| Line 157: | Line 122: | ||
|- | |- | ||
| K<sub>eq</sub> || Equilibrium constant || | | K<sub>eq</sub> || Equilibrium constant || | ||
| + | |- | ||
| + | | X1 , X2 || Variability factors || | ||
|- | |- | ||
| [PGR] || enzyme concentration || μM | | [PGR] || enzyme concentration || μM | ||
| Line 230: | Line 197: | ||
|} | |} | ||
| − | === Extracting Information from | + | === Extracting Information from menthone Production Rates === |
A table will go here | A table will go here | ||
| Line 236: | Line 203: | ||
=== Published Kinetic Parameter Values === | === Published Kinetic Parameter Values === | ||
| − | + | ==== Km Values ==== | |
| − | + | {| | |
| + | ! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 50px;" | Parameter | ||
| + | ! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 100px;" | Directionality | ||
| + | ! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 100px;" | Substrate / Product | ||
| + | ! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 50px;" | Value | ||
| + | ! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 50px;" | unit | ||
| + | ! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 300px;" | Method notes | ||
| + | ! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 10px;" | References | ||
| + | |- | ||
| + | | Km || Forward || pulegone || 2.3 || µM || Gene from peppermint oil gland secretory cell cDNA, expressed in E. coli, optimal pH 5.0, menthone:isomenthone ratio of 55:45 || Ringer2003 | ||
| + | |- | ||
| + | | Km || Forward || pulegone || 2.9 || µM || Gene from peppermint oil gland secretory cell cDNA, expressed in E. coli, optimal pH 5.0, menthone:isomenthone ratio of 55:45, Km 2.3 +/- 0.6 || Ringer2003 | ||
| + | |- | ||
| + | | Km || Forward || NADPH || 6.9 || µM || Gene from peppermint oil gland secretory cell cDNA, expressed in E. coli, optimal pH 5.0, menthone:isomenthone ratio of 55:45 || Ringer2003 | ||
| + | |} | ||
=== Detailed description of kinetic values obtained from literature === | === Detailed description of kinetic values obtained from literature === | ||
| Line 245: | Line 226: | ||
== Simulations == | == Simulations == | ||
| + | The DBR model have been used to simulate the biotransformation experiment from published experiments. Click on the pictures below to learn more: | ||
| + | |||
| + | <imagemap> | ||
| + | File:DBR_biotrans_toogood_simulation.png| 500px | ||
| + | rect 41 49 941 684 [[Simulating DBR Biotransformation]] | ||
| + | desc bottom-left | ||
| + | </imagemap> | ||
== References == | == References == | ||
<references /> | <references /> | ||
Latest revision as of 13:36, 23 March 2017
You can go back to main page of the kinetic model here.
Contents
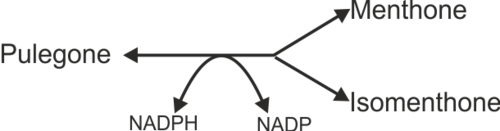
Reaction catalysed
Enzyme and Metabolite Background Information
Long metabolite names are abbreviated in the model for clarity and standard identification purposes.
| Compound | Abbreviation | Chemical Formula | Molar mass (g/mol) | ChEBI | ChEMBL | PubChem | BRENDA | PlantCyc |
|---|---|---|---|---|---|---|---|---|
| Double-bond reductase | DBR | 37914 Da | 1.3.1.81 | |||||
| pulegone | C10H16O | 136.24 | ||||||
| menthone | ||||||||
| NADPH | C21H30N7O17P3 | 745.42116 | 16474 | |||||
| NADP+ | C21H29N7O17P3 | 744.41322 | 18009 | |||||
| isomenthone |
Equation Rate
DBR is modelled using two equations that describes: 1) Pulegone to Menthone and 2) Pulegone to Isomenthone. The predominant product for DBR is menthone where the ratio of menthone to isomenthone production have been reported as 40:33 [1], 55:45 [2] and 70:30 [3]. To model the variability of the product yield for DBR, variability factors which are calculated from the average ratio is included in the equations.
Reaction 1: Conversion of pulegone to menthone
Reaction 2: Conversion of pulegone to isomenthone
| Parameter | Description | Units |
|---|---|---|
| VPGR | Reaction rate for Limonene-3-hydroxylase | μM/min |
| Kcatforward | Catalytic constant in the forward direction | s-1 |
| Kmpulegone | Michaelis-Menten constant for pulegone | μM |
| Kmmenthone | Michaelis-Menten constant for menthone | μM |
| Kmisomenthone | Michaelis-Menten constant for isomenthone | μM |
| KmNADPH | Michaelis-Menten constant for NADPH | μM |
| KmNADP | Michaelis-Menten constant for NADP+ | μM |
| Keq | Equilibrium constant | |
| X1 , X2 | Variability factors | |
| [PGR] | enzyme concentration | μM |
| [pulegone] | Pulegone concentration | μM |
| [menthone] | Menthone concentration | μM |
| [isomenthone] | Isomenthone concentration | μM |
| [NADPH] | NADPH concentration | μM |
| [NADP] | NADP+ concentration | μM |
Strategies for estimating the kinetic parameter values
Standard Gibbs Free energy
The Gibbs free energy for PGR is -3.9565125 kcal.mol^-1. This value is estimated from the 'Contribution group' method by Latendresse, M. and is available from MetaCyc (EC 1.3.1.81) [4].
Calculating the Equilibrium Constant
The equilibrium constant can be calculated using the Van't Hoff Isotherm equation:
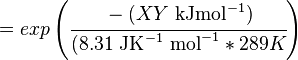
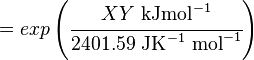
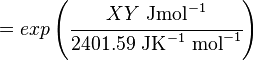
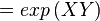
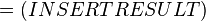
where;
| Keq | Equilibrium constant |
| -ΔG° | Gibbs free energy change. For (INSERT ENZYME) it is (INSERT VALUE) kJmol-1 |
| R | Gas constant with a value of 8.31 JK-1mol-1 |
| T | Temperature which is always expressed in kelvin |
Extracting Information from menthone Production Rates
A table will go here
Published Kinetic Parameter Values
Km Values
| Parameter | Directionality | Substrate / Product | Value | unit | Method notes | References |
|---|---|---|---|---|---|---|
| Km | Forward | pulegone | 2.3 | µM | Gene from peppermint oil gland secretory cell cDNA, expressed in E. coli, optimal pH 5.0, menthone:isomenthone ratio of 55:45 | Ringer2003 |
| Km | Forward | pulegone | 2.9 | µM | Gene from peppermint oil gland secretory cell cDNA, expressed in E. coli, optimal pH 5.0, menthone:isomenthone ratio of 55:45, Km 2.3 +/- 0.6 | Ringer2003 |
| Km | Forward | NADPH | 6.9 | µM | Gene from peppermint oil gland secretory cell cDNA, expressed in E. coli, optimal pH 5.0, menthone:isomenthone ratio of 55:45 | Ringer2003 |
Detailed description of kinetic values obtained from literature
A more detailed description of the values listed above can be found here .
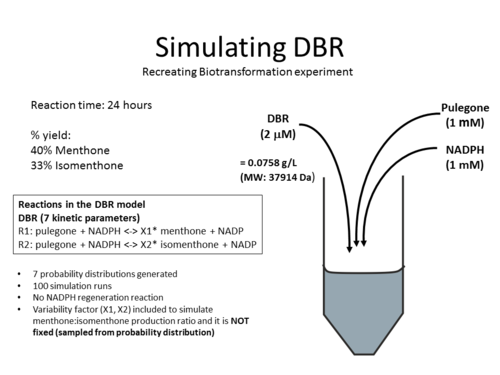
Simulations
The DBR model have been used to simulate the biotransformation experiment from published experiments. Click on the pictures below to learn more:
References
- ↑ 1.0 1.1 Toogood, H. et. al.2015. "Enzymatic menthol production: One-pot approach using engioneered Escherichia coli ", ACS Synthetic Biology, 4:1112-1123 Cite error: Invalid
<ref>tag; name "Toogood2015" defined multiple times with different content - ↑ Ringer, K.L. et. al. 2003. "Monoterpene double-bond reductases of the (-)-menthol biosynthetic pathway:isolation and characterization of cDNAs encoding (-)-isopiperitenone reductase and (+)-pulegone reductase of peppermint", Archives of Biochemistry and Biophysics 418:80-92
- ↑ Solodar, J. 1976. "Stereoselective reduction of menthone and isomenthone by dissolving alkali metals and by hydrogen with Group 8 Metals on carbon", The Journal of Organic Chemistry, 41(21):3461-3464
- ↑ Latendresse M. (2013). "Computing Gibbs Free Energy of Compounds and Reactions in MetaCyc."
![V_\mathrm{DBR} = Kcat_\mathrm{forward} * X1*[DBR] * \cfrac {\left ( \cfrac{[pulegone]}{Km_\mathrm{pulegone}} * \cfrac {[NADPH]}{Km_\mathrm{NADPH}} \right ) * \left ( 1 - \cfrac {[menthone]*[NADP]}{[pulegone]*[NADPH]*K_\mathrm{eq}} \right )}
{ \left (1 + \cfrac {[Pulegone]}{Km_\mathrm{Pulegone}} + \cfrac {[NADP]}{Km_\mathrm{NADP}} \right ) + \left ( 1+ \cfrac {[NADPH]}{Km_\mathrm{NADPH}} + \cfrac {[menthone]}{Km_\mathrm{menthone}} \right ) }](/wiki/images/math/e/8/2/e8266ee0a10232ab7cf83510dde96773.png)
![V_\mathrm{DBR} = Kcat_\mathrm{forward} *X2* [DBR] * \cfrac {\left ( \cfrac{[pulegone]}{Km_\mathrm{pulegone}} * \cfrac {[NADPH]}{Km_\mathrm{NADPH}} \right ) * \left ( 1 - \cfrac {[isomenthone]*[NADP]}{[pulegone]*[NADPH]*K_\mathrm{eq}} \right )}
{ \left (1 + \cfrac {[Pulegone]}{Km_\mathrm{Pulegone}} + \cfrac {[NADP]}{Km_\mathrm{NADP}} \right ) + \left ( 1+ \cfrac {[NADPH]}{Km_\mathrm{NADPH}} + \cfrac {[isomenthone]}{Km_\mathrm{isomenthone}} \right ) }](/wiki/images/math/b/1/f/b1f27f5e860b074806c3b452ce27db1c.png)