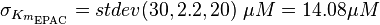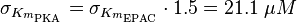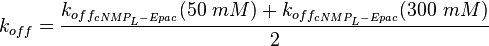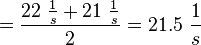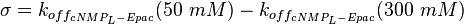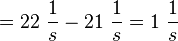Difference between revisions of "EPAC and PKA-Activation (Agonist)"
m |
m |
||
| (8 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{:Template:MAPK_Template}} | {{:Template:MAPK_Template}} | ||
| + | |||
| + | The EPAC and PKA-Activation is modelled in two different ways. Once to keep as close as possible to Xu et al. (2010)<ref name="Xu2010"> Tian-Rui Xu et al. (2010) "Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species." ''Sci Signal''. 3(134):ra20. ([http://www.ncbi.nlm.nih.gov/pubmed/20234003 pmid:20234003])</ref> following Michaelis-Menten kinetics (non-modified model) and once as mass action kinetics (modified model), because the activation of those molecules is not due to an enzymatic catalysation but due to binding of cAMP (Kraemer A. et al. 2001)<ref name="Kraemer2001">Kraemer A. et al. (2001) "Dynamic interaction of cAMP with the Rap guanine-nucleotide exchange factor Epac1." ''J Mol Biol'' 306.5:1167-1177. ([http://www.ncbi.nlm.nih.gov/pubmed/11237625 pmid:11237625])</ref>. | ||
| + | |||
<h2> Binding Reaction </h2> | <h2> Binding Reaction </h2> | ||
<p><br></p> | <p><br></p> | ||
| − | <center><math> inactiveEPAC + agonist \rightleftharpoons activeEPAC + agonist </math></center> | + | <center><math> \text{inactiveEPAC} + \text{agonist} \rightleftharpoons \text{activeEPAC} + \text{agonist} </math></center> |
| − | <center><math> inactivePKA + agonist \rightleftharpoons activePKA + agonist </math></center> | + | <center><math> \text{inactivePKA} + \text{agonist} \rightleftharpoons \text{activePKA} + \text{agonist} </math></center> |
<p><br></p> | <p><br></p> | ||
<h2>Kinetic Equation </h2> | <h2>Kinetic Equation </h2> | ||
<p><br></p> | <p><br></p> | ||
| − | <center><math>\frac{k_{cat} \cdot [agonist] \cdot [inactiveEPAC] \cdot (1-\frac{[activeEPAC]}{[inactiveEPAC] \cdot K_{eq}})}{1 + \frac{[inactiveEPAC]}{Km_{inactiveEPAC}} + \frac{[activeEPAC]}{Km_{activeEPAC}}}</math></center> | + | <center><math>v_{\text{EPAC-Activation}}=\frac{k_{cat} \cdot [\text{agonist}] \cdot [\text{inactiveEPAC}] \cdot (1-\frac{[\text{activeEPAC}]}{[\text{inactiveEPAC}] \cdot K_{eq}})}{Km_{\text{inactiveEPAC}} \cdot (1 + \frac{[\text{inactiveEPAC}]}{Km_{\text{inactiveEPAC}}} + \frac{[\text{activeEPAC}]}{Km_{\text{activeEPAC}}})}</math></center> |
| − | <center><math>\frac{k_{cat} \cdot [agonist] \cdot [inactivePKA] \cdot (1-\frac{[activePKA]}{[inactivePKA] \cdot K_{eq}})}{1 + \frac{[inactivePKA]}{Km_{inactivePKA}} + \frac{[activePKA]}{Km_{activePKA}}}</math></center> | + | <center><math>v_{\text{PKA-Activation}}=\frac{k_{cat} \cdot [\text{agonist}] \cdot [\text{inactivePKA}] \cdot (1-\frac{[\text{activePKA}]}{[\text{inactivePKA}] \cdot K_{eq}})}{Km_{\text{inactiveEPAC}} \cdot (1 + \frac{[\text{inactivePKA}]}{Km_{\text{inactivePKA}}} + \frac{[\text{activePKA}]}{Km_{\text{activePKA}}})}</math></center> |
<p><br></p> | <p><br></p> | ||
| + | |||
<h2>final Parameter </h2> | <h2>final Parameter </h2> | ||
<p><br></p> | <p><br></p> | ||
'''Michaelis-Menten:''' | '''Michaelis-Menten:''' | ||
| − | <math>K_{m_{EPAC} | + | <math>K_{m_{\text{EPAC}}} = 17.4 \pm 14.1\ \mu M </math> |
| − | |||
| − | |||
| − | <math>K_{ | + | <math>K_{m_{\text{PKA}}} = 17.4 \pm 14.1 \cdot 1.5\ \mu M = 17.4 \pm 21.1\ \mu M </math> |
| − | <math>K_{cat_{EPAC}} = 4.07534 | + | <math>K_{cat_{\text{EPAC}}} = 4.07534\ \frac{1}{s} </math> |
| − | <math>K_{cat_{PKA}} = 1.20102 | + | <math>K_{cat_{\text{PKA}}} = 1.20102\ \frac{1}{s} </math> |
| − | |||
<p><br></p> | <p><br></p> | ||
'''Binding reaction''': | '''Binding reaction''': | ||
| − | <math>k_{ | + | <math>k_{on_{\text{EPAC}}} = 15 \pm 5.6\ \frac{1}{\mu M \cdot s}</math> |
| + | |||
| + | <math>k_{off{\text{EPAC}}} = 21.5 \pm 1\ \frac{1}{s}</math> | ||
| + | |||
| + | <math>k_{on_{\text{PKA}}} = 15 \pm 5.6\cdot 1.5 \frac{1}{\mu M \cdot s} = 15 \pm 8.4\ \frac{1}{\mu M \cdot s}</math> | ||
| + | |||
| + | <math>k_{off{\text{PKA}}} = 21.5 \pm 1\cdot 1.5 \frac{1}{s} = 21.5 \pm 1.5\ \frac{1}{s}</math> | ||
| + | |||
| + | <!-- Takeda et al. 2011<ref name="Takeda2011">Takeda S. et al. (2011) "Systems analysis of GLP-1 receptor signaling in pancreatic β-cells." ''Am J Pysiol'' 301.4:C792-C803 ([http://www.ncbi.nlm.nih.gov/pubmed/21734192 pmid:21734192]) </ref> --> | ||
| − | |||
<h2>Parameter Annotations</h2> | <h2>Parameter Annotations</h2> | ||
| − | |||
| − | + | <!-- [[File:L EPAC-Activation.png|thumb|500px|Dao et al. (2006)<ref name="Dao2006"></ref>]]--> | |
| − | < | + | |
| − | [[File:L EPAC-Activation.png| | + | <table border="1"> |
| + | <tr> | ||
| + | <th>Notes</th> | ||
| + | <th>References</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p>To calculate the Km values for EPAC and PKA the average of three measured values is used. The Km value for the activation by cAMP is measured twice and the activation by 8CPT-2Me-cAMP once. Because this reaction is included in our model to increase comparability with Xu et al's model<ref name="Xu2010"></ref> and they used this analogon for their measurements all three values are averaged. </p> | ||
| + | |||
| + | <p> The values are:</p> | ||
| + | |||
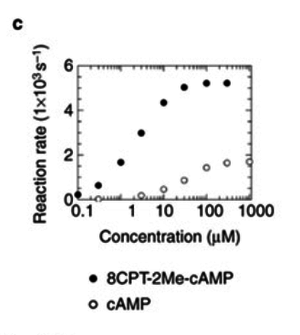
| + | <p>Km<sub>cAMP</sub>(EPAC1) = 30 μM; Enserink et al.(2002)<ref name="Enserink2002"> Enserik J.M. et al. (2002) "A novel Epac-specific cAMP analogue demonstrates independent regulation of Rap1 and ERK." ''Nat Cell Biol.'' 4.11:901-906 ([http://www.ncbi.nlm.nih.gov/pubmed/12402047 pmid:12402047])</ref></p> | ||
| − | < | + | <p> Km<sub>8CPT-2Me-cAMP</sub>(EPAC1) = 2.2 μM; Enserink et al. (2002)<ref name="Enserink2002"></ref></p> |
| − | + | ||
| + | <p> Km<sub>cAMP</sub>(EPAC2) = 20 μM; Rehmann H. et al. (2008)<ref name="Rehmann2008">Rehmann H. et al. (2008) "Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B" ''Nature'' 455,124-127. ([http://www.ncbi.nlm.nih.gov/pubmed/18660803 pmid:18660803]) </ref></p> | ||
| − | + | <p> There are no measured values for the PKA activation by cAMP. However it can be assumed that the values are similar to the ones for EPAC activation. Therefore the same value as for EPAC activation is used and the assumed error is the error used for EPAC activation increased by 50%, because the reaction is not the reaction measured. | |
| + | </p> | ||
| − | + | <p> '''Calculation''' </p> | |
| + | <math> K_{m_{\text{EPAC & PKA}}} = \frac{30\; \mu M + 2.2\; \mu M + 20\; \mu M}{3} = 17.4\; \mu M </math> | ||
| − | + | <math> \sigma_{K_{m_{\text{EPAC}}}} = stdev(30, 2.2, 20)\; \mu M = 14.08 \mu M </math> | |
| − | + | <math> \sigma_{K_{m_{\text{PKA}}}} = \sigma_{K_{m_{\text{EPAC}}}} \cdot 1.5 = 21.1\; \mu M </math> | |
| − | + | </td> | |
| + | <td> | ||
| + | <table> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <div class="thumb tleft"><div class="thumbinner" style="width:300px;"> | ||
| + | [[File:L Km values Enserink.png|border|300px|]][[File:L Km values Enserink1.png|border|300px]]<div class="thumbcaption"> <p align="right"> Enserink et al.(2002)<ref name="Enserink2002"></ref></p></div></div></div> | ||
| + | </td> | ||
| + | <td> | ||
| + | <div class="thumb tright"><div class="thumbinner" style="width:400px;"> | ||
| + | [[File:L Epac2 value Rehmann.png|border|400px|]]<div class="thumbcaption"> <p align="right"> Rehmann H. et al. (2008); Supplementary <ref name="Rehmann2008"></ref></p></div></div></div> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
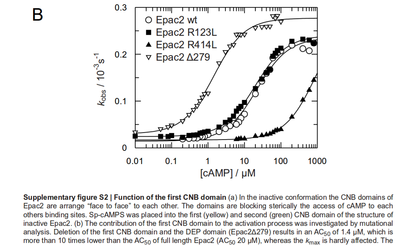
| − | It is shown, that the catalytic domain of EPAC has no influence on the binding ability of | + | The k<sub>cat</sub> values by Xu et al. (2010)<ref name="Xu2010"></ref> are used and to estimate the error the relative errors of all measured k<sub>cat</sub> values used in this model are averaged and this value is assumed to be the relative error of this k<sub>cat</sub> value. ([[Calculation of the averaged relative error]]) |
| − | ). Binding experiment between the cAMP-binding domain (cNMP<sub>L</sub>-EPAC) and cAMP were performed at two different NaCl-level. The dissociation rate differs only slightly, whereas the association rate seems to be dependent on the NaCl-level. | + | </td> |
| + | <td> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | It is shown, that the catalytic domain of EPAC has no influence on the binding ability of EPAC concerning cAMP (Kraemer et al., 2001<ref name="Kraemer2001"></ref>). Binding experiment between the cAMP-binding domain (cNMP<sub>L</sub>-EPAC) and cAMP were performed at two different NaCl-level. The dissociation rate differs only slightly, whereas the association rate seems to be dependent on the NaCl-level. | ||
The intracellular sodium concentration in ''E.coli'' varies between 5-14 mM (Lo et al. 2006<ref name="Lo2006">Lo C., Leake M.C. and Berry R.M. (2006) "Fluorescence measurement of intracellular sodium concentration in single Escherichia coli cells." ''Biophys J.'' 90.1:357-365 ([http://www.ncbi.nlm.nih.gov/pubmed/16227503 pmid:16227503])</ref>) and can therefore be assumed as less than 50 mM. | The intracellular sodium concentration in ''E.coli'' varies between 5-14 mM (Lo et al. 2006<ref name="Lo2006">Lo C., Leake M.C. and Berry R.M. (2006) "Fluorescence measurement of intracellular sodium concentration in single Escherichia coli cells." ''Biophys J.'' 90.1:357-365 ([http://www.ncbi.nlm.nih.gov/pubmed/16227503 pmid:16227503])</ref>) and can therefore be assumed as less than 50 mM. | ||
| Line 64: | Line 108: | ||
Calculation of the k<sub>on</sub>: | Calculation of the k<sub>on</sub>: | ||
| − | Due to the fact that there are only two values measured, no | + | Due to the fact that there are only two values measured, no assumption can be made about the function which describes the relation between the sodium concentration and the association constant. However at higher sodium concentrations a decrease of k<sub>on</sub> was observed and this justifies the assumption that at lower sodium concentrations the association rate might be even higher. It might however be the other way round as well, because possibly the peak of the function is at 50 mM NaCl. We assume that the true value at an intracellular NaCl concentration <50mM is slightly higher than the value measured at 50mM, so the mean of k<sub>on</sub> is set to 15 1/μMs. To estimate the uncertainty of this parameter the difference between the two measured values is set to σ. |
Calculation of the k<sub>off</sub>: | Calculation of the k<sub>off</sub>: | ||
| Line 77: | Line 121: | ||
koff<sub>cNMP_L-Epac</sub> = 21 s<sup>-1</sup>; 300 mM NaCl (Kraemer et al., 2001)<ref name="Kraemer2001"></ref> | koff<sub>cNMP_L-Epac</sub> = 21 s<sup>-1</sup>; 300 mM NaCl (Kraemer et al., 2001)<ref name="Kraemer2001"></ref> | ||
| + | </td> | ||
| + | <td> | ||
| + | <div class="thumb tleft" style="width:80%;"><div class="thumbinner" style="width:100%;"> | ||
| + | [[File: L CAMP to EPAC Binding.png|border|600px|]]<div class="thumbcaption"> <p align="right"> Kraemer et al. (2001)<ref name="Kraemer2001"></ref></p></div></div></div> | ||
| + | <p><br style="clear:left;"/></p> | ||
| + | <p align="left">'''Calculation k<sub>off</sub>:'''</p> | ||
| + | |||
| + | average: | ||
| + | |||
| + | <math>k_{off} = \frac{k_{off_{cNMP_L-Epac}}(50\ mM)+k_{off_{cNMP_L-Epac}}(300\ mM)}{2}</math> | ||
| + | |||
| + | <math>=\frac{22\ \frac{1}{s} + 21\ \frac{1}{s}}{2} = 21.5\ \frac{1}{s}</math> | ||
| + | |||
| + | uncertainty: | ||
| + | |||
| + | <math>\sigma = k_{off_{cNMP_L-Epac}}(50\ mM) - k_{off_{cNMP_L-Epac}}(300\ mM) </math> | ||
| + | |||
| + | <math>= 22\ \frac{1}{s} - 21\ \frac{1}{s} = 1\ \frac{1}{s}</math> | ||
| + | |||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | <h2>Km<sub>P</sub> value and equilibrium constant </h2> | ||
| + | The K<sub>m</sub> value for the product is assumed to be similar but slightly higher than the the K<sub>m</sub> value of the substrate because of the similarity of the both species. Therefore the K<sub>m</sub> value of the substrate is multiplied by 1.05 to gain the one of the product and the uncertainty is increased by increasing the error on Km<sub>substrate</sub> by 50%. | ||
| + | For information about the equilibrium constant please see [[Equilibrium_constants|here]]. | ||
<h2>References</h2> | <h2>References</h2> | ||
<references/> | <references/> | ||
Latest revision as of 11:30, 12 June 2014
The EPAC and PKA-Activation is modelled in two different ways. Once to keep as close as possible to Xu et al. (2010)[1] following Michaelis-Menten kinetics (non-modified model) and once as mass action kinetics (modified model), because the activation of those molecules is not due to an enzymatic catalysation but due to binding of cAMP (Kraemer A. et al. 2001)[2].
Binding Reaction
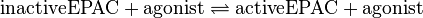
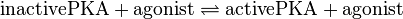
Kinetic Equation
![v_{\text{EPAC-Activation}}=\frac{k_{cat} \cdot [\text{agonist}] \cdot [\text{inactiveEPAC}] \cdot (1-\frac{[\text{activeEPAC}]}{[\text{inactiveEPAC}] \cdot K_{eq}})}{Km_{\text{inactiveEPAC}} \cdot (1 + \frac{[\text{inactiveEPAC}]}{Km_{\text{inactiveEPAC}}} + \frac{[\text{activeEPAC}]}{Km_{\text{activeEPAC}}})}](/wiki/images/math/1/a/2/1a21573904fc04e23cd538672d957fcb.png)
![v_{\text{PKA-Activation}}=\frac{k_{cat} \cdot [\text{agonist}] \cdot [\text{inactivePKA}] \cdot (1-\frac{[\text{activePKA}]}{[\text{inactivePKA}] \cdot K_{eq}})}{Km_{\text{inactiveEPAC}} \cdot (1 + \frac{[\text{inactivePKA}]}{Km_{\text{inactivePKA}}} + \frac{[\text{activePKA}]}{Km_{\text{activePKA}}})}](/wiki/images/math/7/0/2/7029809930067c14104e1caa41a7c178.png)
final Parameter
Michaelis-Menten:
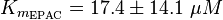
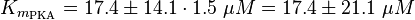
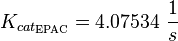
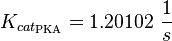
Binding reaction:
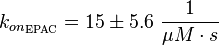
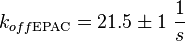
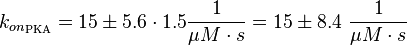
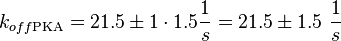
Parameter Annotations
| Notes | References | ||
|---|---|---|---|
|
To calculate the Km values for EPAC and PKA the average of three measured values is used. The Km value for the activation by cAMP is measured twice and the activation by 8CPT-2Me-cAMP once. Because this reaction is included in our model to increase comparability with Xu et al's model[1] and they used this analogon for their measurements all three values are averaged. The values are: KmcAMP(EPAC1) = 30 μM; Enserink et al.(2002)[3] Km8CPT-2Me-cAMP(EPAC1) = 2.2 μM; Enserink et al. (2002)[3] KmcAMP(EPAC2) = 20 μM; Rehmann H. et al. (2008)[4] There are no measured values for the PKA activation by cAMP. However it can be assumed that the values are similar to the ones for EPAC activation. Therefore the same value as for EPAC activation is used and the assumed error is the error used for EPAC activation increased by 50%, because the reaction is not the reaction measured. Calculation Failed to parse (lexing error): K_{m_{\text{EPAC & PKA}}} = \frac{30\; \mu M + 2.2\; \mu M + 20\; \mu M}{3} = 17.4\; \mu M
|
|
||
|
The kcat values by Xu et al. (2010)[1] are used and to estimate the error the relative errors of all measured kcat values used in this model are averaged and this value is assumed to be the relative error of this kcat value. (Calculation of the averaged relative error) |
|||
|
It is shown, that the catalytic domain of EPAC has no influence on the binding ability of EPAC concerning cAMP (Kraemer et al., 2001[2]). Binding experiment between the cAMP-binding domain (cNMPL-EPAC) and cAMP were performed at two different NaCl-level. The dissociation rate differs only slightly, whereas the association rate seems to be dependent on the NaCl-level. The intracellular sodium concentration in E.coli varies between 5-14 mM (Lo et al. 2006[5]) and can therefore be assumed as less than 50 mM. Calculation of the kon: Due to the fact that there are only two values measured, no assumption can be made about the function which describes the relation between the sodium concentration and the association constant. However at higher sodium concentrations a decrease of kon was observed and this justifies the assumption that at lower sodium concentrations the association rate might be even higher. It might however be the other way round as well, because possibly the peak of the function is at 50 mM NaCl. We assume that the true value at an intracellular NaCl concentration <50mM is slightly higher than the value measured at 50mM, so the mean of kon is set to 15 1/μMs. To estimate the uncertainty of this parameter the difference between the two measured values is set to σ. Calculation of the koff: The two values differ only sightly, so we assume that the dissociation rate does not depend on the sodium concentration. Therefore the two values are averaged to get the mean and the error is estimated similar to kon. The measured values are: koncNMP_L-Epac = 14.4 1/μMs; 50 mM NaCl (Kraemer et al., 2001)[2] koncNMP_L-Epac= 8.8 1/μMs; 300 mM NaCl (Kraemer et al., 2001)[2] koffcNMP_L-Epac = 22 s-1; 50 mM NaCl (Kraemer et al., 2001)[2] koffcNMP_L-Epac = 21 s-1; 300 mM NaCl (Kraemer et al., 2001)[2] |
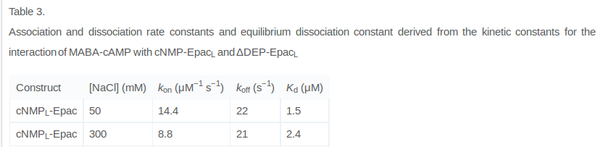 Kraemer et al. (2001)[2] Calculation koff: average:
uncertainty:
|
KmP value and equilibrium constant
The Km value for the product is assumed to be similar but slightly higher than the the Km value of the substrate because of the similarity of the both species. Therefore the Km value of the substrate is multiplied by 1.05 to gain the one of the product and the uncertainty is increased by increasing the error on Kmsubstrate by 50%.
For information about the equilibrium constant please see here.
References
- ↑ 1.0 1.1 1.2 Tian-Rui Xu et al. (2010) "Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species." Sci Signal. 3(134):ra20. (pmid:20234003)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Kraemer A. et al. (2001) "Dynamic interaction of cAMP with the Rap guanine-nucleotide exchange factor Epac1." J Mol Biol 306.5:1167-1177. (pmid:11237625)
- ↑ 3.0 3.1 3.2 Enserik J.M. et al. (2002) "A novel Epac-specific cAMP analogue demonstrates independent regulation of Rap1 and ERK." Nat Cell Biol. 4.11:901-906 (pmid:12402047)
- ↑ 4.0 4.1 Rehmann H. et al. (2008) "Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B" Nature 455,124-127. (pmid:18660803)
- ↑ Lo C., Leake M.C. and Berry R.M. (2006) "Fluorescence measurement of intracellular sodium concentration in single Escherichia coli cells." Biophys J. 90.1:357-365 (pmid:16227503)