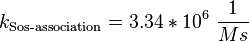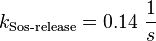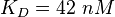Difference between revisions of "Sos-Binding"
(Created page with "<h2> Reaction </h2> <math>inactiveSos + activeEGFR \rightleftharpoons activeSos</math> <h2> Rate Equation </h2> <math>v_{Sos-binding} = k_{Sos-binding} * [activeEGFR][inactiv...") |
|||
| (14 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Template:MAPK_Template}} | ||
| + | |||
| + | |||
<h2> Reaction </h2> | <h2> Reaction </h2> | ||
| − | <math>inactiveSos + activeEGFR \rightleftharpoons activeSos</math> | + | <center><math>\text{inactiveSos} + \text{activeEGFR} \rightleftharpoons \text{activeSos}</math></center> |
| + | <br/> | ||
| + | <h2> Rate Equation </h2> | ||
| + | <center><math>v_{\text{Sos-binding}} = k_{\text{Sos-binding}} \cdot [\text{activeEGFR}][\text{inactiveSos}] - k_{\text{Sos-release}} \cdot [\text{activeSos}]</math></center> | ||
| + | <br/> | ||
| + | <h2>General Information about the Mechanism </h2> | ||
| + | <p> | ||
| + | Sos is a guanine nucleotide release protein (GNRP), which binds via its C-terminal SH3 domain to Grb2 | ||
| + | <ref name="Tanaka1994">Tanaka S. et al. (1994) "C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins" ''Proc. Natl. Acad. Sci. USA'' 91.1:3443-3447([http://www.ncbi.nlm.nih.gov/pubmed/7512734 pmid:7512734])</ref>. | ||
| + | Grb2 has two SH3 domains, but only the N-terminal one is included in the Sos binding process. The binding occures in a 2:1 stoichiometry. The complex then binds via the SH2 domain of Grb2 to the autophosphorylated EGFR. | ||
| + | <ref name="McDonald2012">McDonald C et al. (2012) "Multivalent binding and facilitated diffusion account for the formation of the Grb2-Sos1 signaling complex in a cooperative manner." ''Biochemistry'' 51.10:2122-2135 ([http://www.ncbi.nlm.nih.gov/pubmed/22360309 pmid:22360309])</ref> | ||
| + | <ref name="Ogura2013">Ogura K. and Okamura H. (2013) "Conformational change of Sos-derived proline-rich peptide upon binding Grb2 N-terminal SH3 domain porbed by NMR." ''Sci.Rep.'' 3:2913 ([http://www.ncbi.nlm.nih.gov/pubmed/24105423 pmid:24105423])</ref> | ||
| + | This binding might include a further protein, which is phosphorylated by the active EGFR, Shc. Through to the binding Sos is recruited to the membrane and can therefore activate Ras, the first enzyme of the MAP-kinase. | ||
| + | <ref name="DongChen1996">Dong Chen et al. (1996) "SOS phosphorylation and disassociation of the Grb2-SOS complex by the ERK and JNK signaling pathways." ''J.Biol.Chem.'' 271.11:6328-6332 ([http://www.ncbi.nlm.nih.gov/pubmed/8626428 pmid:8626428])</ref> | ||
| + | <ref name="Smit1996o">Smit L., van der Horst G., and Borst J. (1996) "Sos, Vav, and C3G participate in B-cell receptor-induced signaling pathways and differentially associate with Shc-Grb2, Crk, and Crk-L adaptors." ''J.Biol.Chem.'' 271.15:8564-8569 ([http://www.ncbi.nlm.nih.gov/pubmed/8621483 pmid:8621483])</ref> | ||
| + | |||
| + | Because of this process and further experiments | ||
| + | <ref name="Kumkhaek2013">Kumkhaek et al. (2013) "MASL1 induces erythroid differentiation in human erythropoietin-dependent CD34+ cells through the Raf/MEK/ERK pathway." ''Blood'' 121.16:3216-3227 ([http://www.ncbi.nlm.nih.gov/pubmed/23327923 pmid:23327923])</ref> | ||
| + | <ref name="Freeman2012">Freeman J. et al. (2012) "A high-content imaging workflow to study Grb2 signaling complexes by expression cloning." ''J.Vis.Exp.'' 68. pii:4382 ([http://www.ncbi.nlm.nih.gov/pubmed/23150065 pmid:23150065])</ref> | ||
| + | ,which showed that the activation of Sos is rather through a recruitment to the membrane than to an enzymatic activation we decided to model this activation process as binding, whereas Xu et al modelled it as Michealis-Menten. | ||
| + | <ref name="Xu2010">Xu T. et al. (2010) "Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species." '' Sci.Signal'' 3.134:ra20 ([http://www.ncbi.nlm.nih.gov/pubmed/20731071 pmid:20731071])</ref> | ||
| + | |||
| + | One reason to model the activation as Michaelis-Menten kinetic might be the findings of Ichiba et al, that the negative influence of the termini on the catalytic domain of Sos is repressed by various mechanisms like, phosphorylation, calcium binding or diacylglycerol binding. | ||
| + | <ref name="Ichiba1999">Ichiba et al. (1999) "Activation of C3G guanine nucleotide exchange factor for Rap1 by phosphorylation of tyrosine 504." ''J.Biol.Chem.'' 274.20:14376-14381 ([http://www.ncbi.nlm.nih.gov/pubmed/10318861 pmid:10318861])</ref> | ||
| − | + | </p> | |
| − | |||
<h2> final Parameter </h2> | <h2> final Parameter </h2> | ||
| − | <math>k_{Sos-binding}= 1.47 \cdot 10^8 \pm | + | <p> The errors of the final parameter are increased by 50% because only a similar reaction is measured. </p> |
| − | <math>k_{Sos-release} = 0. | + | <math>k_{\text{Sos-binding}}= 1.47 \cdot 10^8 \pm 4.0873 \cdot 10^7 \cdot 1.5\ \frac{1}{M \cdot s} = 1.47 \cdot 10^2 \pm 6.1310 \cdot 10^1\ \frac{1}{\mu M \cdot s}</math> |
| + | |||
| + | <math>k_{\text{Sos-release}} = 0.1488 \pm 2.643 \cdot 10^{-2} \cdot 1.5\ \frac{1}{s} = 0.1488 \pm 3.965 \cdot 10^{-2}\ \frac{1}{s}</math> | ||
<h2> Parameter </h2> | <h2> Parameter </h2> | ||
| + | |||
| + | <table width=100% border=1px> | ||
| + | <tr> | ||
| + | <th>Notes</th> | ||
| + | <th>References</th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td colspan="2"> | ||
| + | <p> | ||
| + | <math>k_{\text{Sos-association}}= 3.34 * 10^6\ \frac{1}{Ms}</math><br/> | ||
| + | <math>k_{\text{Sos-release}}= 0.14\ \frac{1}{s}</math><br/> | ||
| + | <math>K_D = 42\ nM </math> | ||
| + | </p> | ||
| + | |||
| + | <p>The binding of the p85αSH2-N domain to 17-mer phosphopeptide similar to a PDGFR region. (Hensmann et al. 1994)<ref name="Hensmann1994">Hensmann M. et al. (1994) "Phosphopeptide binding to the N-terminal SH2 domain of the p85 alpha subunit of PI3'-kinase: a heteronuclear NMR study." ''Protein Sci'' 3.7:1020-1030. DOI: 10.1002/pro.5560030704. ([http://www.ncbi.nlm.nih.gov/pubmed/7522724 pmid:7522724]) </ref></p> | ||
| + | |||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td> | ||
| + | <p>The binding of the two different SH2 domains to phosphotyrosine containing peptides derived from an insuline receptor substrate.</p> | ||
| + | </td> | ||
| + | <td> | ||
| + | <p>[[File:L_Sos-Binding1.png|thumb|500px| Felder et al. (1993) <ref name="Felder1993">Felder S. et al. (1993) "SH2 domains exhibit high-affinity binding to tyrosine-phosphorylated peptides yet also exhibit rapid dissociation and exchange." ''Mol Cell Biol'' 13.3:1449-1455 ([http://www.ncbi.nlm.nih.gov/pubmed/7680095 pmid:7680095])</ref>]]</p> | ||
| + | <p></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | <h2>Parameter Calculation</h2> | ||
| + | In general it can be said that there is no direct binding between Sos and the EGFR. "Sos is constitutivly associated with the SH3 domain of Grb2"<ref name="Smit1996o"></ref>, so the binding process modelled here resembles more the binding of the Sos-Grb2 complex to the EGFR. Because of the reason that "Grb2 consists of (only one)[...] SH2 domain"<ref name="Smit1996o"></ref>, only the data derived from the binding of one SH2 domain to the peptide is used for the calculation. | ||
| + | |||
| + | |||
| + | <math>k_{\text{association}}=\frac{1}{5} \cdot (3.34 \cdot 10^6\ \frac{1}{Ms} + 0.48 \cdot 10^8\ \frac{1}{Ms} + 4.4 \cdot 10^8\ \frac{1}{Ms} + 0.33 \cdot 10^8\ \frac{1}{Ms} + 2.1 \cdot 10^8\ \frac{1}{Ms}) = 1.47 \cdot 10^8\ \frac{1}{Ms}</math><br/> | ||
| + | |||
| + | |||
| + | <math>k_{\text{release}}=\frac{1}{5} \cdot (0.14\ \frac{1}{s} + 0.156\ \frac{1}{s} + 0.149\ \frac{1}{s} + 1.113\ \frac{1}{s} + 0.186\ \frac{1}{s}) = 0.1488\ \frac{1}{s} </math> | ||
| + | |||
| + | |||
| + | For the uncertainty estimation of the k<sub>release</sub> the standard deviation of the five values (0.14 1/s, 0.156 1/s, 0.149 1/s, 0.113 1/s, 0.186 1/s) is calculated. | ||
| + | |||
| + | Then the relative error on the mean is calculated <math> \frac{2.643 \cdot 10^{-2}\ \frac{1}{s}}{0.1488\ \frac{1}{s}} = 1.776 \cdot 10^{-1} </math>. This value is assumed to be the relative error of each single measurement. | ||
| + | Taking the measured K<sub>D</sub> error into account the relative error on each single k<sub>on</sub> value can be calculated. To finally estimate the uncertainty on k<sub>on</sub> the average of these four values is taken and assumed as relative error on the calculated k<sub>association</sub> value. | ||
| + | |||
| + | |||
| + | <math>\sigma_{k_{\text{association}}}\frac{0.1226 + 0.3224 + 0.1782 + 0.489}{4} \cdot 1.47 \cdot 10^8 \frac{1}{M \cdot s} = 4.0873 \frac{1}{M \cdot s}</math> | ||
| + | |||
| + | The value measured by (Hensmann et al. 1994)<ref name="Hensmann1994"></ref> is not included in the uncertainty estimation. | ||
| + | |||
| + | |||
| + | <table border="1"> | ||
| + | <tr> | ||
| + | <th style="padding:2px">K<sub>D</sub> value [nM]</th> | ||
| + | <th style="padding:2px">error K<sub>D</sub> [nM]</th> | ||
| + | <th style="padding:2px">relative error K<sub>D</sub></th> | ||
| + | <th style="padding:2px">error calculation k<sub>on</sub></th> | ||
| + | <th style="padding:2px">relative error k<sub>on</sub> </th> | ||
| + | <tr> | ||
| + | <td style="padding:2px"><math>3.27</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>0.18</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>\frac{0.18}{3.27} = 5.5 \cdot 10^{-2}</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math> 5.5 \cdot 10^{-2} - 1.776 \cdot 10^{-1}</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>1.226 \cdot 10^{-1}</math> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="padding:2px"><math>0.34</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>0.17</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>\frac{0.17}{0.34} = 5.5 </math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>5.5 - 1.776 \cdot 10^{-1}</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>3.224 \cdot 10^{-1}</math> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="padding:2px"><math>3.40</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>1.21</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>\frac{1.21}{3.40} = 3.56 \cdot 10^{-1}</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>3.56 \cdot 10^{-1} - 1.776 \cdot 10^{-1}</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>1.782 \cdot 10^{-1}</math> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="padding:2px"><math>0.90</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>0.60</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>\frac{0.60}{0.90} = 6.67 \cdot 10^{-1}</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>6.67 \cdot 10^{-1} - 1.776 \cdot 10^{-1}</math> | ||
| + | </td> | ||
| + | <td style="padding:2px"><math>4.890 \cdot 10^{-1}</math> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h2>References</h2> | ||
| + | <references/> | ||
Latest revision as of 10:41, 31 March 2014
Reaction
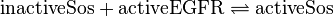
Rate Equation
![v_{\text{Sos-binding}} = k_{\text{Sos-binding}} \cdot [\text{activeEGFR}][\text{inactiveSos}] - k_{\text{Sos-release}} \cdot [\text{activeSos}]](/wiki/images/math/9/e/8/9e89443bf768b7fc49d815f34b4dc7c6.png)
General Information about the Mechanism
Sos is a guanine nucleotide release protein (GNRP), which binds via its C-terminal SH3 domain to Grb2 [1]. Grb2 has two SH3 domains, but only the N-terminal one is included in the Sos binding process. The binding occures in a 2:1 stoichiometry. The complex then binds via the SH2 domain of Grb2 to the autophosphorylated EGFR. [2] [3] This binding might include a further protein, which is phosphorylated by the active EGFR, Shc. Through to the binding Sos is recruited to the membrane and can therefore activate Ras, the first enzyme of the MAP-kinase. [4] [5] Because of this process and further experiments [6] [7] ,which showed that the activation of Sos is rather through a recruitment to the membrane than to an enzymatic activation we decided to model this activation process as binding, whereas Xu et al modelled it as Michealis-Menten. [8] One reason to model the activation as Michaelis-Menten kinetic might be the findings of Ichiba et al, that the negative influence of the termini on the catalytic domain of Sos is repressed by various mechanisms like, phosphorylation, calcium binding or diacylglycerol binding. [9]
final Parameter
The errors of the final parameter are increased by 50% because only a similar reaction is measured.
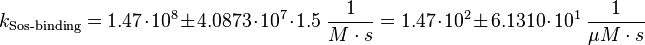
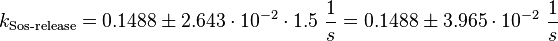
Parameter
| Notes | References |
|---|---|
|
The binding of the p85αSH2-N domain to 17-mer phosphopeptide similar to a PDGFR region. (Hensmann et al. 1994)[10] |
|
|
The binding of the two different SH2 domains to phosphotyrosine containing peptides derived from an insuline receptor substrate. |
 Felder et al. (1993) [11] |
Parameter Calculation
In general it can be said that there is no direct binding between Sos and the EGFR. "Sos is constitutivly associated with the SH3 domain of Grb2"[5], so the binding process modelled here resembles more the binding of the Sos-Grb2 complex to the EGFR. Because of the reason that "Grb2 consists of (only one)[...] SH2 domain"[5], only the data derived from the binding of one SH2 domain to the peptide is used for the calculation.
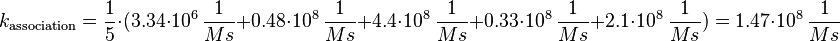
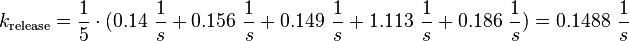
For the uncertainty estimation of the krelease the standard deviation of the five values (0.14 1/s, 0.156 1/s, 0.149 1/s, 0.113 1/s, 0.186 1/s) is calculated.
Then the relative error on the mean is calculated 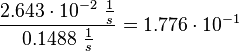 . This value is assumed to be the relative error of each single measurement.
Taking the measured KD error into account the relative error on each single kon value can be calculated. To finally estimate the uncertainty on kon the average of these four values is taken and assumed as relative error on the calculated kassociation value.
. This value is assumed to be the relative error of each single measurement.
Taking the measured KD error into account the relative error on each single kon value can be calculated. To finally estimate the uncertainty on kon the average of these four values is taken and assumed as relative error on the calculated kassociation value.
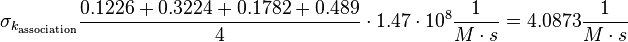
The value measured by (Hensmann et al. 1994)[10] is not included in the uncertainty estimation.
| KD value [nM] | error KD [nM] | relative error KD | error calculation kon | relative error kon |
|---|---|---|---|---|
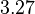
|
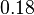
|
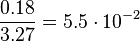
|
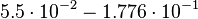
|
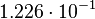
|
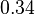
|
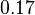
|
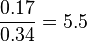
|
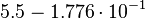
|
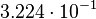
|
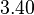
|
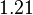
|
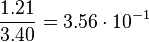
|
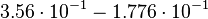
|
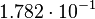
|
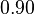
|
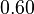
|
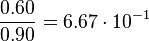
|
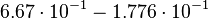
|
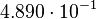
|
References
- ↑ Tanaka S. et al. (1994) "C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins" Proc. Natl. Acad. Sci. USA 91.1:3443-3447(pmid:7512734)
- ↑ McDonald C et al. (2012) "Multivalent binding and facilitated diffusion account for the formation of the Grb2-Sos1 signaling complex in a cooperative manner." Biochemistry 51.10:2122-2135 (pmid:22360309)
- ↑ Ogura K. and Okamura H. (2013) "Conformational change of Sos-derived proline-rich peptide upon binding Grb2 N-terminal SH3 domain porbed by NMR." Sci.Rep. 3:2913 (pmid:24105423)
- ↑ Dong Chen et al. (1996) "SOS phosphorylation and disassociation of the Grb2-SOS complex by the ERK and JNK signaling pathways." J.Biol.Chem. 271.11:6328-6332 (pmid:8626428)
- ↑ 5.0 5.1 5.2 Smit L., van der Horst G., and Borst J. (1996) "Sos, Vav, and C3G participate in B-cell receptor-induced signaling pathways and differentially associate with Shc-Grb2, Crk, and Crk-L adaptors." J.Biol.Chem. 271.15:8564-8569 (pmid:8621483)
- ↑ Kumkhaek et al. (2013) "MASL1 induces erythroid differentiation in human erythropoietin-dependent CD34+ cells through the Raf/MEK/ERK pathway." Blood 121.16:3216-3227 (pmid:23327923)
- ↑ Freeman J. et al. (2012) "A high-content imaging workflow to study Grb2 signaling complexes by expression cloning." J.Vis.Exp. 68. pii:4382 (pmid:23150065)
- ↑ Xu T. et al. (2010) "Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species." Sci.Signal 3.134:ra20 (pmid:20731071)
- ↑ Ichiba et al. (1999) "Activation of C3G guanine nucleotide exchange factor for Rap1 by phosphorylation of tyrosine 504." J.Biol.Chem. 274.20:14376-14381 (pmid:10318861)
- ↑ 10.0 10.1 Hensmann M. et al. (1994) "Phosphopeptide binding to the N-terminal SH2 domain of the p85 alpha subunit of PI3'-kinase: a heteronuclear NMR study." Protein Sci 3.7:1020-1030. DOI: 10.1002/pro.5560030704. (pmid:7522724)
- ↑ Felder S. et al. (1993) "SH2 domains exhibit high-affinity binding to tyrosine-phosphorylated peptides yet also exhibit rapid dissociation and exchange." Mol Cell Biol 13.3:1449-1455 (pmid:7680095)